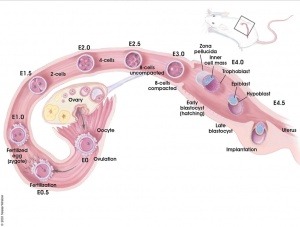
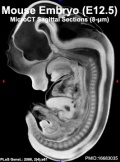
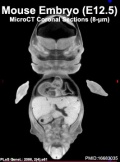
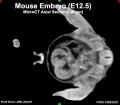
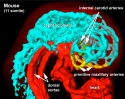
Mouse Development
| Embryology - 7 Mar 2026 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
The mouse (taxon-mus) has always been a good embryological model, generating easily (litters 8-20) and quickly (21d). Mouse embryology really expanded when molecular biologists used mice for gene knockouts. Suddenly it was necessary to understand development in order to understand the effect of knocking out the gene.
There are over 450 different strains of inbred research mice, and these strains have recently been organized into a chart. The deer mouse (Peromyscus maniculatus)[1] lives in the Americas and is closely related to the white-footed mouse. Those interested in the mouse reproductive cycle should also look at the mouse estrous cycle.
There are several systems for staging mouse development. The original and most widely used is the Theiler Stages system, which divides mouse development into 26 prenatal and 2 postnatal stages.[2]
| Mouse Links: Introduction | Mouse Stages | Mouse Timeline | Mouse Timeline Detailed | Mouse Estrous Cycle | Mouse Heart | Mouse Knockout | Movie - Cephalic Plexus | Movie - Blastocyst Cdx2 | ANAT2341 Project 2009 | Category:Mouse | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
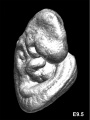
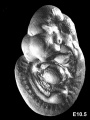
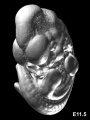
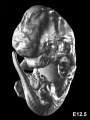
- Mouse Stages: E1 | E2.5 | E3.0 | E3.5 | E4.5 | E5.0 | E5.5 | E6.0 | E7.0 | E7.5 | E8.0 | E8.5 | E9.0 | E9.5 | E10 | E10.5 | E11 | E11.5 | E12 | E12.5 | E13 | E13.5 | E14 | E14.5 | E15 | E15.5 | E16 | E16.5 | E17 | E17.5 | E18 | E18.5 | E19 | E20 | Timeline | About timed pregnancy
| Carnegie | Stage | |||||||||||||||||||||||
| Human | Days | 1 | 2-3 | 4-5 | 5-6 | 7-12 | 13-15 | 15-17 | 17-19 | 20 | 22 | 24 | 28 | 30 | 33 | 36 | 40 | 42 | 44 | 48 | 52 | 54 | 55 | 58 |
| Mouse | Days | 1 | 2 | 3 | E4.5 | E5.0 | E6.0 | E7.0 | E8.0 | E9.0 | E9.5 | E10 | E10.5 | E11 | E11.5 | E12 | E12.5 | E13 | E13.5 | E14 | E14.5 | E15 | E15.5 | E16 |
| Rat | Days | 1 | 3.5 | 4-5 | 5 | 6 | 7.5 | 8.5 | 9 | 10.5 | 11 | 11.5 | 12 | 12.5 | 13 | 13.5 | 14 | 14.5 | 15 | 15.5 | 16 | 16.5 | 17 | 17.5 |
| Note these Carnegie stages are only approximate day timings for average of embryos. Links: Carnegie Stage Comparison | ||||||||||||||||||||||||
| ||||||||||||||||||||||||
| Timeline Links: human timeline | mouse timeline | mouse detailed timeline | chicken timeline | rat timeline | Medaka | Category:Timeline |
Some Recent Findings
<html5media width="550" height="360">https://www.youtube.com/embed/P4kP0Eg348I</html5media>
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Mouse Development |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Movies
The collapsible tables below contain a number of movies showing aspects of mouse development.
| Mouse Zygote | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
| |||||||||||||||
| Mouse Various | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
| |||||||||||||||
|
|
|
|
| |||||||||||||||
| Mouse microCT | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
| |||||||||||||||
|
|
|
|
| |||||||||||||||
- Links: Movies
Animal Model Comparison
| Postnatal Animal Models | mouse | rat | pig |
|---|---|---|---|
| Pregnancy period (days) | 18 – 21 | 21 – 23 | 110 – 118 |
| Placenta type | Discoidal, decidual hemoendothelial choroidea |
Discoidal, decidual hemoendothelial choroidea |
Epitheliochorial |
| Litter size | 6 – 12 | 6 – 15 | 11 – 16 |
| Birth weight (g) | 0.5 – 1.5 | 3 – 5 | 900 – 1600 |
| Weaning weight male/female (g) | 18 – 25/16 – 25 | 55 – 90/45 – 80 | 6000 – 8000 |
| Suckling period (days) | 21–28 | 21 | 28–49 |
| Solid diet beginning (days) | 10 | 12 | 12 – 15 |
| Puberty male/female (week) | 4 – 6/5 | 6/6 – 8 | 20 – 28 |
| Life expectancy (years) | 1 - 2 | 2 - 3 | 14 – 18 |
| Table data - Otis and Brent (1954)[16] Links: timeline | |||
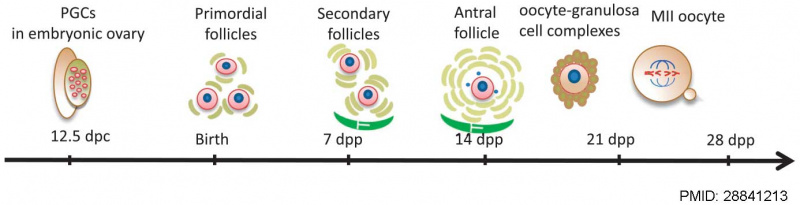
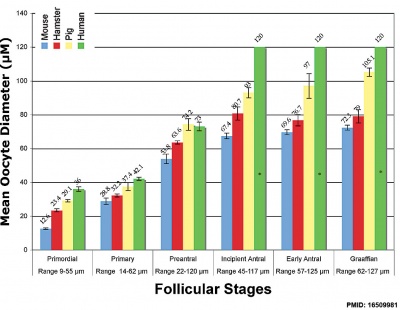
Oocyte Development
The Mose oocyte reaches a maximum size of 70 μm in diameter in a follicle 125 μm in diameter, followed by ongoing follicle growth theca interns and antrum development.<ref name=Brambell1928{{Ref-Brambell1928}}</ref>
Mouse oogenesis [17]
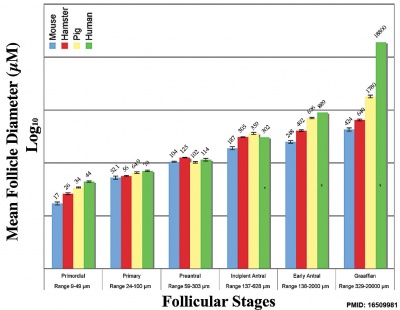
Graphs show mouse and other species (Human, Pig, Hamster) comparison in follicle and oocyte size growth.[18] (See also Follicle size graph)
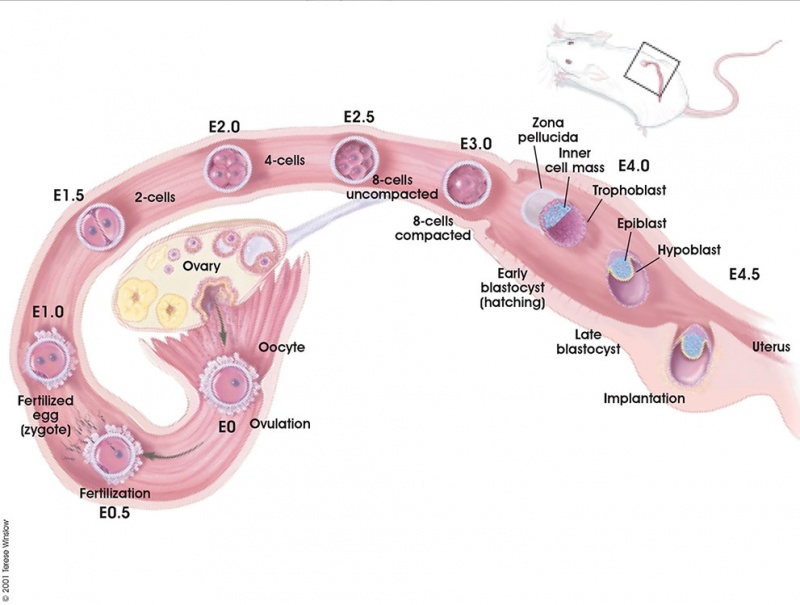
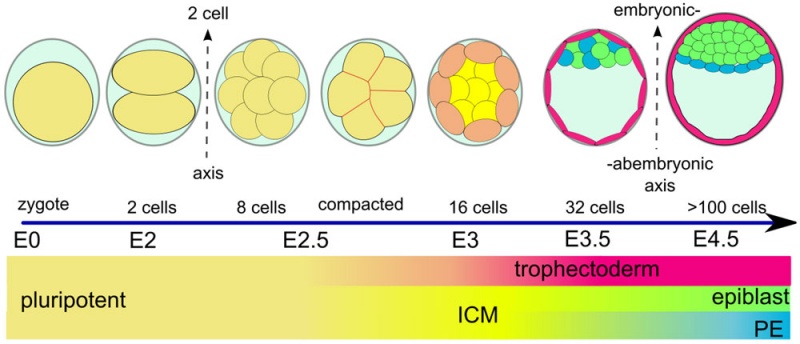
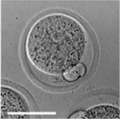
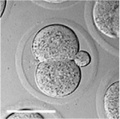
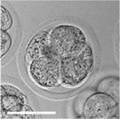
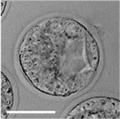
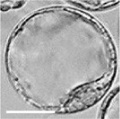
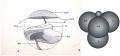
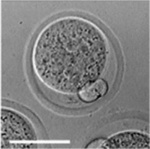
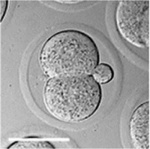
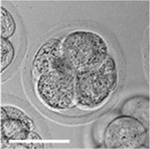
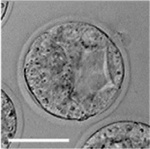
Early Mouse Development
|
|
|
|
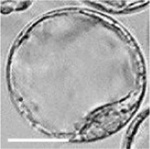
|
| zygote | blastomeres | morula | early blastocyst | hatched blastocyst |
Early mouse development model[19]
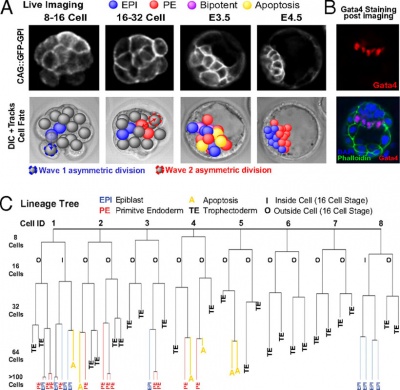
|
|||||||||
Live cell imaging and tracking[20]
|
Simulation of Zygote to Blastocyst[19]
|
- Links: Mouse Stages
Later Mouse Development
- Links: Mouse Stages | Mouse Timeline Detailed
Mouse Limb
- Links: Limb Development
Carnegie Stages Comparison
The table below gives an approximate comparison of human, mouse and rat embryos based upon Carnegie staging.
| Carnegie | Stage | |||||||||||||||||||||||
| Human | Days | 1 | 2-3 | 4-5 | 5-6 | 7-12 | 13-15 | 15-17 | 17-19 | 20 | 22 | 24 | 28 | 30 | 33 | 36 | 40 | 42 | 44 | 48 | 52 | 54 | 55 | 58 |
| Mouse | Days | 1 | 2 | 3 | E4.5 | E5.0 | E6.0 | E7.0 | E8.0 | E9.0 | E9.5 | E10 | E10.5 | E11 | E11.5 | E12 | E12.5 | E13 | E13.5 | E14 | E14.5 | E15 | E15.5 | E16 |
| Rat | Days | 1 | 3.5 | 4-5 | 5 | 6 | 7.5 | 8.5 | 9 | 10.5 | 11 | 11.5 | 12 | 12.5 | 13 | 13.5 | 14 | 14.5 | 15 | 15.5 | 16 | 16.5 | 17 | 17.5 |
| Note these Carnegie stages are only approximate day timings for average of embryos. Links: Carnegie Stage Comparison | ||||||||||||||||||||||||
| ||||||||||||||||||||||||
Placenta Development
Maternal
- Uterine radial artery branches into 5-10 dilated spiral arteries.[21]
- Spiral arteries then converge at the trophoblast giant cell layer and empty into straight trophoblast-lined "canals".
Embryonic
- Embryonic placenta originates from the ectoplacental cone and the extra-embryonic ectoderm.
- endothelial cells derive from the allantois.
- embryonic day (E 10) - placenta divided into three layers associated with maternal decidual cells.
- Labyrinth - (equivalent to human villi) a selective barrier on the fetal side, is an array of fetal and maternal vessels.
- Junctional zone - (spongy layer) produce hormones and contains numerous cavities. The trophoblast cells form spongiotrophoblasts and the glycogen cells, that later (E 12.5) migrate into the maternal decidua.
- Giant cells - next to the uterine cells form the outermost fetal cell layer until (E 12).
| Mouse Placenta Vasculature (E16.5) | |
|---|---|
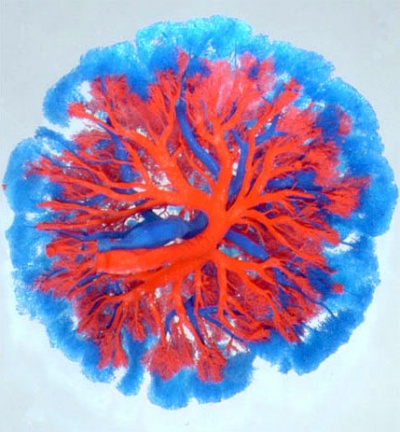
|

|
| superior view (maternal side) | lateral view (maternal side at bottom) |
Spermatozoa Development
The process of spermatogenesis takes approximately 35 days:
- mitotic phase (11 days)
- meiotic phase (10 days)
- post-meiotic phase (14 days)
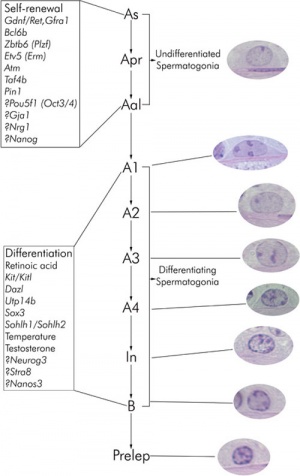
Spermatogonial stem cells (SSCs)
The diploid germ cells, spermatogonial stem cells (SSCs), are located on the basement membrane of the seminiferous tubules
- adult mouse testis about 30,000 SSCs
- either divides into two new single cells
- or into a pair of spermatogonia (Apr)
- that do not complete cytokinesis and stay connected by an intercellular bridge
Primitive spermatogonia subset
- Asingle (As, single isolated spermatogonia)
- Apaired (Apr, interconnected spermatogonial pairs)
- Aaligned (Aal, interconnected 4, 8, or 16 spermatogonia)
- specifically termed Aal-4, Aal-8, and Aal-16
Primitive spermatogonia cells transform without cell division into more differentiating A1 spermatogonia that undergo 6 mitotic and 2 meiotic divisions to eventually form haploid spermatids.
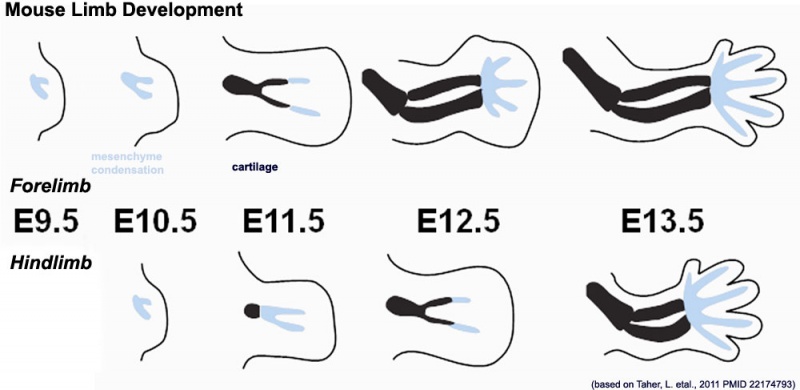
Limb Development
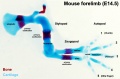
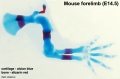
Mouse limb skeleton cartoon[22]
Fore-limb and hind-limb buds for stages E9.5 to E13.5. Hindlimbs are morphologically delayed by about half a day.
- Light blue - indicate mesenchymal condensations.
- Thick black lines - indicate cartilage as determined by alcian blue staining.
Change in cell types and tissue formation as a function of mouse developmental stage.[22]
- Links: Limb Development
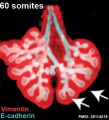
Neural Development
The adult mouse brain consist of approximately 70 million neurons.[23]
Early Neural Development
Neural Fold Fusion[24]
- first fusion - at the level of the intermediate point between the third and fourth somites (caudal myelencephalon) both rostrally and caudally.
- second fusion - at the original rostral end of the neural plate (rostrodorsally).
- third fusion - in the caudal diencephalon (rostrally and caudally)
- followed by complete closure of the telencephalic neuropore at the midpoint of the telencephalic roof
- then complete closure of the metencephalic neuropore at the rostral part of the metencephalic roof
- fourth fusion - at the original caudal end of the neural plate (rostrally)
- caudal neuropore completely closed at the level of the future 33rd somite
See also these 1980's papers: neural plate cells of early-somite-stage mouse embryos[25] and Neurulation in the mouse.[26]
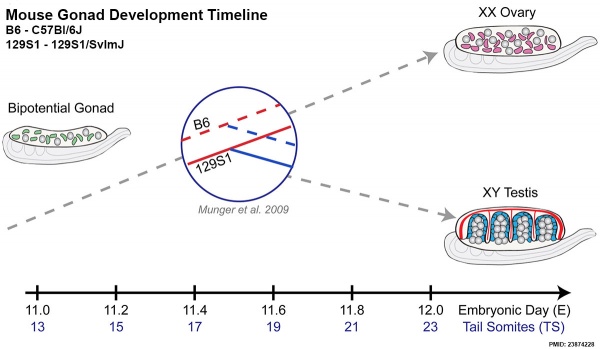
Urogenital Development
Mouse E11.0 to E12.0 shows the critical transition in the gonad from a bipotential to sexually-differentiated state. Based upon 2013 transcriptome analysis.[27]
A high-resolution description of the developing murine genitourinary tract from Theiler stage (TS) 17 (E10.5) through to TS27 (E19.5) and then to postnatal day 3 was published in 2007.[28]
The GenitoUrinary Development Molecular Anatomy Project (GUDMAP) is a consortium of laboratories working to provide the scientific and medical community with tools to facilitate research. They intend to develop:
- a molecular atlas of gene expression for the developing organs of the GenitoUrinary (GU) tract
- a high resolution molecular anatomy that highlights development of the GU system
- mouse strains to facilitate developmental and functional studies within the GU system
- tutorials describing GU organogenesis
- rapid access to primary data via the GUDMAP database
Lung Development
Vertebrate lung development is generally divided into 5 stages, based upon growth and histological appearance. Mouse age data[29]
| Stage | Mouse Age | Features |
|---|---|---|
| Embryonic | E9 to E11.5 | lung buds originate as an outgrowth from the ventral wall of the foregut where lobar division occurs |
| Pseudoglandular | E11.5 to E16.5 | conducting epithelial tubes surrounded by thick mesenchyme are formed, extensive airway branching |
| Canalicular | E16.5 to E17.5 | bronchioles are produced, increasing number of capillaries in close contact with cuboidal epithelium and the beginning of alveolar epithelium development |
| Saccular | E17.5 to PN5 | alveolar ducts and air sacs are developed |
| Alveolar | PN5 to PN28 | secondary septation occurs, marked increase of the number and size of capillaries and alveoli |
Human and Mouse Comparison
| Stage | Human | Mouse | Features |
|---|---|---|---|
| Embryonic | week 4 to 5 | E9 to E11.5 | lung buds originate as an outgrowth from the ventral wall of the foregut where lobar division occurs |
| Pseudoglandular | week 5 to 17 | E11.5 to E16.5 | conducting epithelial tubes surrounded by thick mesenchyme are formed, extensive airway branching |
| Canalicular | week 16 to 25 | E16.5 to E17.5 | bronchioles are produced, increasing number of capillaries in close contact with cuboidal epithelium and the beginning of alveolar epithelium development |
| Saccular | week 24 to 40 | E17.5 to PN5 | alveolar ducts and air sacs are developed |
| Alveolar | late fetal to 8 years | PN5 to PN28 | secondary septation occurs, marked increase of the number and size of capillaries and alveoli |
Note that while human third trimester fetus has regular respiratory breathing movements, the common mouse model (C57BL/6J} completely lacks these movements.[30]
Respiratory Species Comparison
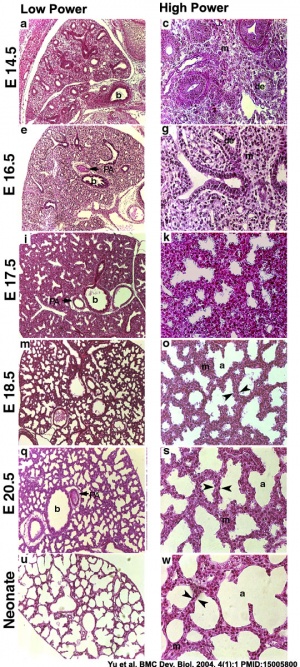
| Respiratory Stages - Species Comparison - Stages Gestational age (days) | |||||
|---|---|---|---|---|---|
| Species | Term | Embryonic | Pseudoglandular | Canalicular | Saccular |
| human | 280 | < 42 | 52 - 112 | 112 - 168 | 168 |
| primate | 168 | < 42 | 57 - 80 | 80 - 140 | 140 |
| sheep | 150 | < 40 | 40 - 80 | 80 - 120 | 120 |
| rabbit | 32 | < 18 | 21 - 24 | 24 - 27 | 27 |
| rat | 22 | < 13 | 16 - 19 | 19 - 20 | 21 |
| mouse | 20 | < 9 | 16 | 18 | 19 |
| Data modified from[32]
Links: respiratory | Respiratory Comparison | Mouse Human Respiratory | Mouse respiratory stages | mouse | rat | rabbit | Timeline Comparisons | |||||
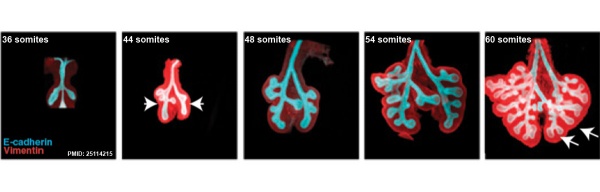
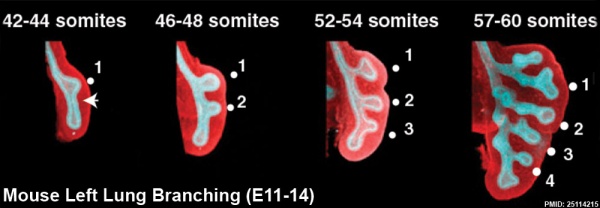
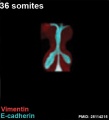
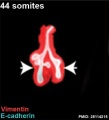
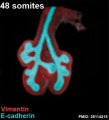
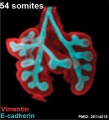
Bronchial Branching
The following images are from a recent study of the development of bronchial branching in he mouse between E10 to E14.[33]
Mesenchyme (red) and epithelium (blue) the study used knockout mice to show the role of Wnt signalling in branching morphogenesis.
Endocrine Development
Hypothalamic-Pituitary-Adrenal Axis
Two postnatal phases identified[34]:
- first phase - (pnd 1 (birth) to pnd 12) corresponds to the hypo-responsive period (SHRP) in the rat. Basal corticosterone levels were low and novelty exposure did not enhance corticosterone or ACTH levels. High expression of CRH in the paraventricular nucleus (PVN) of the hypothalamus. Expression levels of GR in the hippocampus and UCN3 in the perifornical area are low at birth but increase significantly during the SHRP, both reach maximum expression level at pnd 12.
- second phase - mice developed past the SHRP exhibit enhanced corticosterone basal levels and a response of ACTH and corticosterone to mild novelty stress. CRH expression was decreased significantly, expression of urocortin 3 (UCN3) and glucocorticoid receptor (GR) remained high, with a small decrease at pnd 16.
Mouse Knockouts
Knowledge about mouse development has rapidly expanded as it has become the model animal system for genetic "knock out " studies. This technology actually requires development of defined breeding programs, pseudo-pregnancy, in vitro fertilization, molecular biology, and good old fashioned histology. Without understanding normal development the molecular biologists don't stand a hope of understanding what their gene knock out has done. There is a database of all existing mouse knockouts and their consequences.
Murine Development Control Genes
Kessel, M. and Gruss, P. Science 249 374-379 (1990)
An early review of the genes, and method of identifying them, involved in early mouse development. In particular discusses Homeobox genes. (homeobox is 183bp encoding a 61 amino acid DNA-binding domain)
- Gene families
- Hox
- Pax
- POU
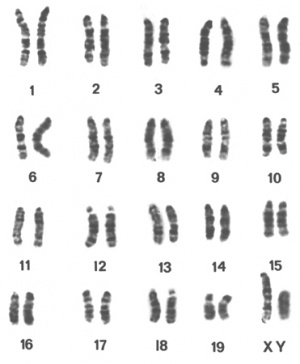
The Genealogy Chart of Inbred Strains
This Nature paper[35] chart shows the origins and relationships of inbred mouse strains. The chart is available as a PDF document [Media:Mouse_genealogy.pdf Locally] or from JAX Labs and was originally published by Beck etal., 2000.
- Hairless mutant mouse - wave of hair loss in a rostral to caudal sequence starting at 3 weeks of age, first identified in the 1930's.
Mouse Genome
Mouse Genome completed December 2002, a draft sequence and analysis of the genome of the C57BL/6J mouse strain.
- less than 30,000 genes
- estimated size is 2.5 Gb, smaller than the human genome
- about 40% of the human and mouse genomes can be directly aligned
- about 80% of human genes have one corresponding gene in the mouse genome
- Links: Mouse Genome Sequencing: Mus musculus | Mouse Genome Informatics | Mouse Genome Project | Nature - Mouse Genome
Animal Model Differences
pancreas - "In contrast to the published data from mouse embryos, during human pancreas development, we detected only a single-phase of Neurogenin 3 (NEUROG3) expression and endocrine differentiation from approximately 8 weeks, before which Nirenberg and Kim homeobox 2.2 (NKX2.2) was not observed in the pancreatic progenitor cell population. In addition to revealing a number of disparities in timing between human and mouse development, these data, directly assembled from human tissue."[36]
White pulp - Human white pulp has three stromal cell types of different phenotype and location.[37]
Marginal zone - only in mouse and rat between white and red pulp an additional B-cell compartment.[37]
Red pulp - mouse and human red pulp sinusoid circulation differs.[38]
- mouse - circulation is closed, afferent arterial blood to the red pulp sinusoids, and to the marginal zone sinus.
- humans - circulation is open, no connection from capillaries to sinuses in the red pulp.
Contrasting mechanisms of penile urethral formation in mouse and human[39] "This paper addresses the developmental mechanisms of formation of the mouse and human penile urethra and the possibility that two disparate mechanisms are at play. It has been suggested that the entire penile urethra of the mouse forms via direct canalization of the endodermal urethral plate. While this mechanism surely accounts for development of the proximal portion of the mouse penile urethra, we suggest that the distal portion of the mouse penile urethra forms via a series of epithelial fusion events. Through review of the recent literature in combination with new data, it is unlikely that the entire mouse urethra is formed from the endodermal urethral plate due in part to the fact that from E14 onward the urethral plate is not present in the distal aspect of the genital tubercle. Formation of the distal portion of the mouse urethra receives substantial contribution from the preputial swellings that form the preputial-urethral groove and subsequently the preputial-urethral canal, the later of which is subdivided by a fusion event to form the distal portion of the mouse penile urethra. Examination of human penile development also reveals comparable dual morphogenetic mechanisms. However, in the case of human, direct canalization of the urethral plate occurs in the glans, while fusion events are involved in formation of the urethra within the penile shaft, a pattern exactly opposite to that of the mouse. The highest incidence of hypospadias in humans occurs at the junction of these two different developmental mechanisms. The relevance of the mouse as a model of human hypospadias is discussed."
References
- ↑ Davis SW & Keisler JL. (2016). Embryonic Development of the Deer Mouse, Peromyscus maniculatus. PLoS ONE , 11, e0150598. PMID: 26930071 DOI.
- ↑ Theiler K. The House Mouse: Atlas of Mouse Development (1972, 1989) Springer-Verlag, NY. Online
- ↑ 3.0 3.1 Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nürnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dollé P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G & Ballabio A. (2011). A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. , 9, e1000582. PMID: 21267068 DOI.
- ↑ Aguilera-Castrejon A & Hanna JH. (2021). Ex Utero Culture of Mouse Embryos from Pregastrulation to Advanced Organogenesis. J Vis Exp , , . PMID: 34747408 DOI.
- ↑ Gerovska D & Araúzo-Bravo MJ. (2019). Computational analysis of single-cell transcriptomics data elucidates the stabilization of Oct4 expression in the E3.25 mouse preimplantation embryo. Sci Rep , 9, 8930. PMID: 31222057 DOI.
- ↑ Chan MM, Smith ZD, Grosswendt S, Kretzmer H, Norman TM, Adamson B, Jost M, Quinn JJ, Yang D, Jones MG, Khodaverdian A, Yosef N, Meissner A & Weissman JS. (2019). Molecular recording of mammalian embryogenesis. Nature , 570, 77-82. PMID: 31086336 DOI.
- ↑ McDole K. etal., In Toto Imaging and Reconstruction of Post-Implantation Mouse Development at the Single-Cell Level DOI:https://doi.org/10.1016/j.cell.2018.09.031
- ↑ 8.0 8.1 Chen VS, Morrison JP, Southwell MF, Foley JF, Bolon B & Elmore SA. (2017). Histology Atlas of the Developing Prenatal and Postnatal Mouse Central Nervous System, with Emphasis on Prenatal Days E7.5 to E18.5. Toxicol Pathol , 45, 705-744. PMID: 28891434 DOI.
- ↑ Lindström NO, McMahon JA, Guo J, Tran T, Guo Q, Rutledge E, Parvez RK, Saribekyan G, Schuler RE, Liao C, Kim AD, Abdelhalim A, Ruffins SW, Thornton ME, Baskin L, Grubbs B, Kesselman C & McMahon AP. (2018). Conserved and Divergent Features of Human and Mouse Kidney Organogenesis. J. Am. Soc. Nephrol. , 29, 785-805. PMID: 29449453 DOI.
- ↑ Crawford LW, Foley JF & Elmore SA. (2010). Histology atlas of the developing mouse hepatobiliary system with emphasis on embryonic days 9.5-18.5. Toxicol Pathol , 38, 872-906. PMID: 20805319 DOI.
- ↑ Swartley OM, Foley JF, Livingston DP, Cullen JM & Elmore SA. (2016). Histology Atlas of the Developing Mouse Hepatobiliary Hemolymphatic Vascular System with Emphasis on Embryonic Days 11.5-18.5 and Early Postnatal Development. Toxicol Pathol , 44, 705-25. PMID: 26961180 DOI.
- ↑ Savolainen SM, Foley JF & Elmore SA. (2009). Histology atlas of the developing mouse heart with emphasis on E11.5 to E18.5. Toxicol Pathol , 37, 395-414. PMID: 19359541 DOI.
- ↑ Nishimura YV, Shinoda T, Inaguma Y, Ito H & Nagata K. (2012). Application of in utero electroporation and live imaging in the analyses of neuronal migration during mouse brain development. Med Mol Morphol , 45, 1-6. PMID: 22431177 DOI.
- ↑ Takaoka K & Hamada H. (2012). Cell fate decisions and axis determination in the early mouse embryo. Development , 139, 3-14. PMID: 22147950 DOI.
- ↑ Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF & Bradley A. (2011). A conditional knockout resource for the genome-wide study of mouse gene function. Nature , 474, 337-42. PMID: 21677750 DOI.
- ↑ Otis EM and Brent R. Equivalent ages in mouse and human embryos. (1954) Anat Rec. 120(1):33-63. PMID 13207763
- ↑ Wang JJ, Ge W, Liu JC, Klinger FG, Dyce PW, De Felici M & Shen W. (2017). Complete in vitro oogenesis: retrospects and prospects. Cell Death Differ. , 24, 1845-1852. PMID: 28841213 DOI.
- ↑ Griffin J, Emery BR, Huang I, Peterson CM & Carrell DT. (2006). Comparative analysis of follicle morphology and oocyte diameter in four mammalian species (mouse, hamster, pig, and human). J. Exp. Clin. Assist. Reprod. , 3, 2. PMID: 16509981 DOI.
- ↑ 19.0 19.1 Krupinski P, Chickarmane V & Peterson C. (2011). Simulating the mammalian blastocyst--molecular and mechanical interactions pattern the embryo. PLoS Comput. Biol. , 7, e1001128. PMID: 21573197 DOI.
- ↑ Morris SA, Teo RT, Li H, Robson P, Glover DM & Zernicka-Goetz M. (2010). Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc. Natl. Acad. Sci. U.S.A. , 107, 6364-9. PMID: 20308546 DOI.
- ↑ Adamson SL, Lu Y, Whiteley KJ, Holmyard D, Hemberger M, Pfarrer C & Cross JC. (2002). Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev. Biol. , 250, 358-73. PMID: 12376109
- ↑ 22.0 22.1 Taher L, Collette NM, Murugesh D, Maxwell E, Ovcharenko I & Loots GG. (2011). Global gene expression analysis of murine limb development. PLoS ONE , 6, e28358. PMID: 22174793 DOI.
- ↑ Herculano-Houzel S, Mota B & Lent R. (2006). Cellular scaling rules for rodent brains. Proc. Natl. Acad. Sci. U.S.A. , 103, 12138-43. PMID: 16880386 DOI.
- ↑ Sakai Y. (1989). Neurulation in the mouse: manner and timing of neural tube closure. Anat. Rec. , 223, 194-203. PMID: 2712345 DOI.
- ↑ Chan WY & Tam PP. (1986). The histogenetic potential of neural plate cells of early-somite-stage mouse embryos. J Embryol Exp Morphol , 96, 183-93. PMID: 3805982
- ↑ Sakai Y. (1987). Neurulation in the mouse. I. The ontogenesis of neural segments and the determination of topographical regions in a central nervous system. Anat. Rec. , 218, 450-7. PMID: 3662046 DOI.
- ↑ Munger SC, Natarajan A, Looger LL, Ohler U & Capel B. (2013). Fine time course expression analysis identifies cascades of activation and repression and maps a putative regulator of mammalian sex determination. PLoS Genet. , 9, e1003630. PMID: 23874228 DOI.
- ↑ Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, Clements D, Cullen-McEwen L, Fleming J, Gilbert T, Herzlinger D, Houghton D, Kaufman MH, Kleymenova E, Koopman PA, Lewis AG, McMahon AP, Mendelsohn CL, Mitchell EK, Rumballe BA, Sweeney DE, Valerius MT, Yamada G, Yang Y & Yu J. (2007). A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr. Patterns , 7, 680-99. PMID: 17452023 DOI.
- ↑ Maeda Y, Davé V & Whitsett JA. (2007). Transcriptional control of lung morphogenesis. Physiol. Rev. , 87, 219-44. PMID: 17237346 DOI.
- ↑ Kleven GA & Ronca AE. (2009). Prenatal behavior of the C57BL/6J mouse: a promising model for human fetal movement during early to mid-gestation. Dev Psychobiol , 51, 84-94. PMID: 18980217 DOI.
- ↑ Yu H, Wessels A, Chen J, Phelps AL, Oatis J, Tint GS & Patel SB. (2004). Late gestational lung hypoplasia in a mouse model of the Smith-Lemli-Opitz syndrome. BMC Dev. Biol. , 4, 1. PMID: 15005800 DOI.
- ↑ Pinkerton KE & Joad JP. (2000). The mammalian respiratory system and critical windows of exposure for children's health. Environ. Health Perspect. , 108 Suppl 3, 457-62. PMID: 10852845
- ↑ Kadzik RS, Cohen ED, Morley MP, Stewart KM, Lu MM & Morrisey EE. (2014). Wnt ligand/Frizzled 2 receptor signaling regulates tube shape and branch-point formation in the lung through control of epithelial cell shape. Proc. Natl. Acad. Sci. U.S.A. , 111, 12444-9. PMID: 25114215 DOI.
- ↑ Schmidt MV, Schmidt M, Enthoven L, van der Mark M, Levine S, de Kloet ER & Oitzl MS. (2003). The postnatal development of the hypothalamic-pituitary-adrenal axis in the mouse. Int. J. Dev. Neurosci. , 21, 125-32. PMID: 12711350
- ↑ Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF & Fisher EM. (2000). Genealogies of mouse inbred strains. Nat. Genet. , 24, 23-5. PMID: 10615122 DOI.
- ↑ Jennings RE, Berry AA, Kirkwood-Wilson R, Roberts NA, Hearn T, Salisbury RJ, Blaylock J, Piper Hanley K & Hanley NA. (2013). Development of the human pancreas from foregut to endocrine commitment. Diabetes , 62, 3514-22. PMID: 23630303 DOI.
- ↑ 37.0 37.1 Steiniger BS. (2015). Human spleen microanatomy: why mice do not suffice. Immunology , 145, 334-46. PMID: 25827019 DOI.
- ↑ Steiniger B, Bette M & Schwarzbach H. (2011). The open microcirculation in human spleens: a three-dimensional approach. J. Histochem. Cytochem. , 59, 639-48. PMID: 21525186 DOI.
- ↑ Liu G, Liu X, Shen J, Sinclair A, Baskin L & Cunha GR. (2018). Contrasting mechanisms of penile urethral formation in mouse and human. Differentiation , 101, 46-64. PMID: 29859371 DOI.
Reviews
Sutherland AE. (2016). Tissue morphodynamics shaping the early mouse embryo. Semin. Cell Dev. Biol. , 55, 89-98. PMID: 26820524 DOI.
Rossant J. (2016). Making the Mouse Blastocyst: Past, Present, and Future. Curr. Top. Dev. Biol. , 117, 275-88. PMID: 26969983 DOI.
Swartley OM, Foley JF, Livingston DP, Cullen JM & Elmore SA. (2016). Histology Atlas of the Developing Mouse Hepatobiliary Hemolymphatic Vascular System with Emphasis on Embryonic Days 11.5-18.5 and Early Postnatal Development. Toxicol Pathol , 44, 705-25. PMID: 26961180 DOI.
Savolainen SM, Foley JF & Elmore SA. (2009). Histology atlas of the developing mouse heart with emphasis on E11.5 to E18.5. Toxicol Pathol , 37, 395-414. PMID: 19359541 DOI.
Articles
Search Pubmed
Search Pubmed: Mouse Development | Mouse Embryology
Additional Images
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
Mouse Embryology
- The e-Mouse Atlas Project EMAP
Mouse Gene Expression
- EMAGE is a database of in situ gene expression data in the mouse embryo and an accompanying suite of tools to search and analyse the data. (All site content, except where otherwise noted, is licensed under a Creative Commons Attribution License.)
- Eurexpress transcriptome atlas a High-Resolution Anatomical Atlas of the Transcriptome in the Mouse Embryo.
- GenePaint.org is a digital atlas of gene expression patterns in the mouse.
- Mouse Imaging Centre
- Gude, W. D. (2012). Histological atlas of the laboratory mouse. Retrieved from https://ebookcentral.proquest.com
Mouse Genome
- NCBI- Mouse Genome Sequencing
- Mouse Genome Informatics
- EBI- Mouse Genome Sequencing
- ANEX Rat and Mouse Database - (University of Tokushima)
- Genetic and Physical Maps of the Mouse Genome (Whitehead Institute/MIT)
- Genetic Location Database (University of Southampton)
- Koop Human and Mouse Genome Project Research (University of Victoria)
- Genome Systems, Inc.
Mouse Jackson Laboratory
- Jackson Laboratory
- Jackson Laboratory Induced Mutant Resource
- Jackson Laboratory BioInformatics Electronic Bulletin Board System
- Mouse Inbred Strains (Jackson Labs)
- Mouse Linkage Map
- Mouse Nomenclature Rules and Guidelines (Jackson Labs)
Mouse Diseases
- Helicobacter hepaticus, a Recently Recognized Bacterial Pathogen, Associated with Chronic Hepatitis and Hepatocellular Neoplasia in Laboratory Mice (CDC)
- Helicobacter Infection in Laboratory Mice: History, Significance, Detection and Management (Charles River)
- Infectious Diseases of Mice and Rats (NAS-ILAR)
- Infectious Diseases of Mice and Rats Companion Guide (NAS-ILAR)
- Infectious Diseases of Mice and Rats (AFIP-POLA)
- Murine Helicobacters (NCI)
Mouse Transgenics
- NIH List of Available Knockout Mice | Factsheet
- International Knockout Mouse Consortium IKMC | Research using IKMC products
- NC3Rs Mouse databases
- Mammary Transgene Database (Baylor College of Medicine)
- TBASE - Transgenic/Targeted Mutation Database
- B Universal Ltd.
- BIG BLUE (Big Blue Transgenic Mouse Mailing List Archives)
- Big Blue lacI Transgenic Mouse Page
- Charles River Laboratories, Inc.
- Chrysalis DNX Transgenic Services
Mouse Transgenic Facilities
- Transgenic Animal Facility (University of Connecticut Biotechnology Center)
- Transgenic Animal Facility (University of Illinois Biotechnology Center)
- Transgenic Animal Facility (University of Iowa)
- Transgenic Animal Facility (Karolinska Institute)
- Transgenic Animal Facility (University of Western Australia)
- Transgenic Animal Facility (University of Wisconsin Biotechnology Center)
- Transgenic Animal Model Core (University of Michigan)
- Transgenic Core (Baylor College of Medicine SPORE)
- Transgenic Facility (University of Colorado Health Science Center)
- Transgenic Facility (University of Kentucky)
- Transgenic & Knockout Mouse Facility (University of Maryland at Baltimore)
- Transgenic Models of Skin Disease Core (Harvard Skin Disease Research Center)
- Transgenic Mouse Core Facilities (Mount Sinai School of Medicine)
- Transgenic Mouse Core Facility (University of Virginia)
- Transgenic Mouse Facility (Columbia-Presbyterian Cancer Center)
- Transgenic Mouse Facility (Duke Comprehensive Cancer Center)
- Transgenic Mouse Facility (SUNY-Stony Brook)
- Transgenic Mouse Facility (University of California-Irvine)
- Transgenic Mouse Facility (University of South Carolina)
Mouse Urogenital
- GenitoUrinary Development Molecular Anatomy Project (GUDMAP) Renal Development Tutorial | Genital Development Tutorial
Mouse Unsorted Links
- Covance Research Products
- Edison Biotechnology Institute (Ohio University)
- Encyclopedia of the Mouse Genome (Jackson Laboratory)
- EUCIB Mouse Backcross Database (MBx)
- Harlan Sprague Dawley, Inc.
- It's a Knockout
- Jackson Laboratory Gopher Server
- Lane List of Named Mutations and Polymorphic Loci
- Lexicon Genetics, Inc.
- Lymphocytic Choriomeningitis Virus: An Unrecognized Teratogenic Pathogen (CDC)
- M & B Breeding and Research Centre Ltd.
- Map Manager Software
- Mice and Men: Making the Most of Our Similarities (ORNL)
- Microinjection Workshop
- Mouse Atlas and Gene Expression Database Project
- Mouse Cytogenetic Map Image
- Mouse Idiograms
- Mouse Linkage Map
- Mouse and Rat Research Home Page
- MouseUp HyperCard Transgenic Mouse Database (Macintosh)
- MRC Mouse Genome Centre
- National Institute of Genetics Mouse Genetic Resources (Japan)
- NIH Animal Genetic Resource
- NIH Directory of Transgenic Mice
- NIH Mouse Club
- Of Mice and Men (HHMI's Blazing a Genetic Trail)
- Online Mendelian Inheritance in Animals (University of Sydney)
- Pathology of Genetically-altered Mice
- Portable Dictionary of the Mouse Genome
- Review of Pasteurella pneumotropica (Charles River)
- Taconic
- The Mouse in Science: Cancer Research
- The Mouse in Science: Monoclonal Antibodies
- The Mouse in Science: Vaccines
- The Mouse in Science: Why Mice?
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2026, March 7) Embryology Mouse Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Mouse_Development
- © Dr Mark Hill 2026, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G