Medaka Development
| Embryology - 27 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
Medaka Oryzias latipes or Japanese rice fish is a member of the killifish family first described in 1846 and has been widely used as a aquarium fish. A modified aquarium version with a genetically modified fluorescent (GFP) version also now available in some countries.
A 2004 study by Iwamatsu[1] has characterised the stages of normal fish development.
Medaka fish were also the first for the first vertebrate animal to mate in space (The International Microgravity Laboratory IML-2/STS-65 mission in 1994) as a developmental model for space experiments. The fish has also been used in studies of pigmentation development.
| Fish Links: Zebrafish Development | Medaka Development | Salmon Development | Movie - Zebrafish Heart | Student Group Project - Zebrafish | Recent References | Category:Zebrafish | Category:Medaka |
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Medaka Embryology |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Taxon
cellular organisms; Eukaryota; Fungi/Metazoa group; Metazoa; Eumetazoa; Bilateria; Coelomata; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Elopocephala; Clupeocephala; Euteleostei; Neognathi; Neoteleostei; Eurypterygii; Ctenosquamata; Acanthomorpha; Euacanthomorpha; Holacanthopterygii; Acanthopterygii; Euacanthopterygii; Percomorpha; Smegmamorpha; Atherinomorpha; Beloniformes; Adrianichthyoidei; Adrianichthyidae; Oryziinae; Oryzias
Development Overview
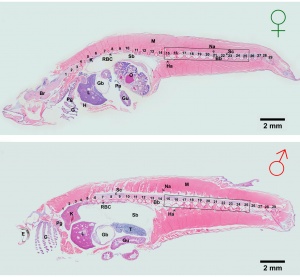
Development has been characterised by light microscope observation into 39 prehatch stages and 6 posthatch stages.[1]
Prehatch features observed included: number and size of blastomeres, form of the blastoderm, extent of epiboly, central nervous system, number and form of somites, optic and otic, notochord, heart, blood circulation, the size and movement of the body, tail, membranous fin (fin fold), viscera (liver gallbladder, gut tube), spleen and swim (air) bladder.
Posthatch features observed included: fins, scales and secondary sexual characteristics.
Developmental Stages
| Stage | Time | Event |
| 0 | Unfertilized eggs | |
| 1 | 3 min | Activated egg |
| 2 | Blastodisc | |
| 3 | 1 h 5 min | 2 cell |
| 4 | 1 h 45 min | 4 cell |
| 5 | 2 h 20 min | 8 cell |
| 6 | 2 h 55 min | 16 cell |
| 7 | 3 h 30 min | 32 cell |
| 8 | 4 h 5 min | Early morula |
| 9 | 5 h 15 min | Late morula |
| 10 | 6 h 30 min | Early blastula |
| 11 | 8 h 15 min | Late blastula |
| 12 | 10 h 20 min | Pre-early gastrula |
| 13 | 13 h | Early gastrula |
| 14 | 15 h | Pre-mid-gastrula |
| 15 | 17 h 30 min | Mid-gastrula |
| 16 | 21 h | Late gastrula |
| 17 | 1 day 1 h | Early neurula (head formation) |
| 18 | 1 day 2 h | Late neurula (optic bud formation) |
| 19 | 1 day 3 h 30 min | 2 somite |
| 20 | 1 day 7 h 30 min | 4 somite |
| 21 | 1 day 10 h | 6 somite (brain regionalization and otic vesicle formation) |
| 22 | 1 day 14 h | 9 somite (appearance of heart anlage) |
| 23 | 1 day 17 h | 12 somite (formation of tubular heart) |
| 24 | 1 day 20 h | 16 somite (start of heart beating) |
| 25 | 2 days 2 h | 18–19 somite (onset of blood circulation) |
| 26 | 2 days 6 h | 22 somite (development of guanophores and vacuolization of the notochord) |
| 27 | 2 days 10 h | 24 somite (appearance of pectoral fin bud) |
| 28 | 2 days 16 h | 30 somite (onset of retinal pigmentation) |
| 29 | 3 days 2 h | 34 somite (internal ear formation) |
| 30 | 3 days 10 h | 35 somite (blood vessel development) |
| 31 | 3 days 23 h | Gill blood vessel formation |
| 32 | 4 days 5 h | Somite completion (formation of pronephros and air bladder) |
| 33 | 4 days 10 h | at which notochord vacuolization is completed |
| 34 | 5 days 1 h | Pectoral fin blood circulation |
| 35 | 5 days 12 h | at which visceral blood vessels form |
| 36 | 6 days | Heart development |
| 37 | 7 days | Pericardial cavity formation |
| 38 | 8 days | Spleen development (differentiation of caudal fin begins) |
| 39 | 9 days | Hatching |
| 40 | 1st fry | |
| 41 | ||
| 42 | ||
| 43 | ||
| 44 | ||
| 45 |
National BioResource Project Medaka
The National BioResource Project Medaka (NBRP Medaka): an integrated bioresource for biological and biomedical sciences[10] "The Japanese government has supported the development of Medaka Bioresources since 2002. The second term of the Medaka Bioresource Project started in 2007. The National Institute for Basic Biology and Niigata University were selected as the core organizations for this project. More than 400 strains including more than 300 spontaneous and induced mutants, 8 inbred lines, 21 transgenic lines, 20 medaka-related species and 66 wild stock lines of medaka are now being provided to the scientific community and educational non-profit organizations. In addition to these live fish, NBRP Medaka is also able to provide cDNA/EST clones such as full-length cDNA and BAC/fosmid clones covering 90% of the medaka genome."
References
- ↑ 1.0 1.1 Iwamatsu T. (2004). Stages of normal development in the medaka Oryzias latipes. Mech. Dev. , 121, 605-18. PMID: 15210170 DOI.
- ↑ Soukup V, Mrstakova S & Kozmik Z. (2018). Asymmetric pitx2 expression in medaka epithalamus is regulated by nodal signaling through an intronic enhancer. Dev. Genes Evol. , 228, 131-139. PMID: 29663064 DOI.
- ↑ Inoue T, Iida A, Maegawa S, Sehara-Fujisawa A & Kinoshita M. (2016). Generation of a transgenic medaka (Oryzias latipes) strain for visualization of nuclear dynamics in early developmental stages. Dev. Growth Differ. , 58, 679-687. PMID: 27759163 DOI.
- ↑ Nishimura T, Sato T, Yamamoto Y, Watakabe I, Ohkawa Y, Suyama M, Kobayashi S & Tanaka M. (2015). Sex determination. foxl3 is a germ cell-intrinsic factor involved in sperm-egg fate decision in medaka. Science , 349, 328-31. PMID: 26067255 DOI.
- ↑ Porazinski S, Wang H, Asaoka Y, Behrndt M, Miyamoto T, Morita H, Hata S, Sasaki T, Krens SFG, Osada Y, Asaka S, Momoi A, Linton S, Miesfeld JB, Link BA, Senga T, Shimizu N, Nagase H, Matsuura S, Bagby S, Kondoh H, Nishina H, Heisenberg CP & Furutani-Seiki M. (2015). YAP is essential for tissue tension to ensure vertebrate 3D body shape. Nature , 521, 217-221. PMID: 25778702 DOI.
- ↑ Rosemore BJ & Welsh CA. (2012). The effects of rearing density, salt concentration, and incubation temperature on Japanese medaka (Oryzias latipes) embryo development. Zebrafish , 9, 185-90. PMID: 23244689 DOI.
- ↑ Renn J & Winkler C. (2009). Osterix-mCherry transgenic medaka for in vivo imaging of bone formation. Dev. Dyn. , 238, 241-8. PMID: 19097055 DOI.
- ↑ Bajoghli B, Aghaallaei N, Jung G & Czerny T. (2009). Induction of otic structures by canonical Wnt signalling in medaka. Dev. Genes Evol. , 219, 391-8. PMID: 19760182 DOI.
- ↑ PLoS One.
- ↑ Sasado T, Tanaka M, Kobayashi K, Sato T, Sakaizumi M & Naruse K. (2010). The National BioResource Project Medaka (NBRP Medaka): an integrated bioresource for biological and biomedical sciences. Exp. Anim. , 59, 13-23. PMID: 20224166
Articles
Liedtke D, Erhard I & Schartl M. (2011). snail gene expression in the medaka, Oryzias latipes. Gene Expr. Patterns , 11, 181-9. PMID: 21094700 DOI.
Shima A & Mitani H. (2004). Medaka as a research organism: past, present and future. Mech. Dev. , 121, 599-604. PMID: 15210169 DOI.
Search Pubmed
Search Pubmed: Medaka Development
External Links
- National BioResource Project Medaka (NBRP Medaka) http://shigen.lab.nig.ac.jp/medaka/
Animal Development
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 27) Embryology Medaka Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Medaka_Development
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G

