Introduction
Amphibian oocyte transcription
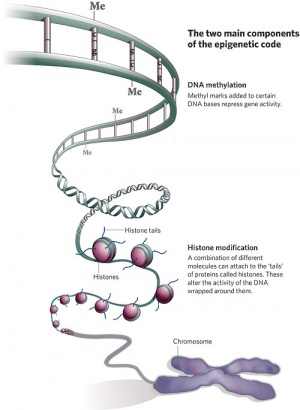
Epigenetics mechanisms
[1]This page is a link to many different resources related to molecular development. I am 2.85 billion nucleotides of DNA, but so is a chimpanzee, and all this DNA encodes only about 20,000-25,000 protein-coding genes. In development, I am not that different from a mouse or a fly and many of the signals that regulate development are used time and time again.
We have come a long way from just observing development to now wanting to understand how the complex program of development is controlled. Using new research tools and some excellent animal models researchers have discovered common themes and mechanisms that tie all embryonic development together.
What is remarkable, given our biological diversity, is the strong evolutionary conservation of developmental mechanisms. This has been a boon in allowing the use of many (easier) model systems such as the genetist's tool the fruitfly, and the worm, frog, chicken, zebrafish and mouse (see Animal Development page).
Gene Regulatory Network (GRN) is a set of genes, or parts of genes, that interact with each other to control a specific cell function. In the embryo, these are the molecular mechanisms/pathways used in the development and differentiation of specific cell types and tissues.
A continuing theme also seems to be the reuse of signals at different times and places within the embryo, for diiferent jobs. This has given rise to the concept of "switches" which by themselves may contain no "information" but to activate other genes or switches. Finally, you can imagine that of our 20,000-25,000 protein-coding genes, a large number of these may only be expressed during development or if reused, have a completely different role in the mature animal. The new field of epigenetics has also begun to florish with some recent important findings.
Molecular mechanisms of development is an exciting area and requires a variety of different skills. This page introduces only a few examples and should give you a feel for the topic. Note that each section of system notes has a page covering molecular development in that system. I have included information about the basic building blocks of proteins (amino acids).
| Hox Genes
|
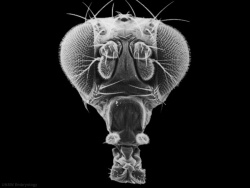
|

|
| Fly wild-type head[2]
|
Fly antennapedia mutant head[2]
|
- Molecular System Links: Heart | Neural | Renal | Respiratory | Mechanisms | Factors | Molecular
Some Recent Findings
- Multimodal transcriptional control of pattern formation in embryonic development[3] "Predicting how interactions between transcription factors and regulatory DNA sequence dictate rates of transcription and, ultimately, drive developmental outcomes remains an open challenge in physical biology. Using stripe 2 of the even-skipped gene in Drosophila embryos as a case study, we dissect the regulatory forces underpinning a key step along the developmental decision-making cascade: the generation of cytoplasmic mRNA patterns via the control of transcription in individual cells. Using live imaging and computational approaches, we found that the transcriptional burst frequency is modulated across the stripe to control the mRNA production rate. However, we discovered that bursting alone cannot quantitatively recapitulate the formation of the stripe and that control of the window of time over which each nucleus transcribes even-skipped plays a critical role in stripe formation. Theoretical modeling revealed that these regulatory strategies (bursting and the time window) respond in different ways to input transcription factor concentrations, suggesting that the stripe is shaped by the interplay of 2 distinct underlying molecular processes." fly
- Key role for CTCF in establishing chromatin structure in human embryos[4] "In the interphase of the cell cycle, chromatin is arranged in a hierarchical structure within the nucleus1,2, which has an important role in regulating gene expression3-6. However, the dynamics of 3D chromatin structure during human embryogenesis remains unknown. Here we report that, unlike mouse sperm, human sperm cells do not express the chromatin regulator CTCF and their chromatin does not contain topologically associating domains (TADs). Following human fertilization, TAD structure is gradually established during embryonic development. In addition, A/B compartmentalization is lost in human embryos at the 2-cell stage and is re-established during embryogenesis. Notably, blocking zygotic genome activation (ZGA) can inhibit TAD establishment in human embryos but not in mouse or Drosophila. Of note, CTCF is expressed at very low levels before ZGA, and is then highly expressed at the ZGA stage when TADs are observed. TAD organization is significantly reduced in CTCF knockdown embryos, suggesting that TAD establishment during ZGA in human embryos requires CTCF expression. Our results indicate that CTCF has a key role in the establishment of 3D chromatin structure during human embryogenesis."
- Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2-1[5] "The extraordinarily thin alveolar type 1 (AT1) cell constitutes nearly the entire gas exchange surface and allows passive diffusion of oxygen into the blood stream. Despite such an essential role, the transcriptional network controlling AT1 cells remains unclear. Using cell-specific knockout mouse models, genomic profiling, and 3D imaging, we found that NK homeobox 2-1 (Nkx2-1) is expressed in AT1 cells and is required for the development and maintenance of AT1 cells. Without Nkx2-1, developing AT1 cells lose 3 defining features-molecular markers, expansive morphology, and cellular quiescence-leading to alveolar simplification and lethality. NKX2-1 is also cell-autonomously required for the same 3 defining features in mature AT1 cells. Intriguingly, Nkx2-1 mutant AT1 cells activate gastrointestinal (GI) genes and form dense microvilli-like structures apically. Single-cell RNA-seq supports a linear transformation of Nkx2-1 mutant AT1 cells toward a GI fate. Whole lung ChIP-seq shows NKX2-1 binding to 68% of genes that are down-regulated upon Nkx2-1 deletion, including 93% of known AT1 genes, but near-background binding to up-regulated genes. Our results place NKX2-1 at the top of the AT1 cell transcriptional hierarchy and demonstrate remarkable plasticity of an otherwise terminally differentiated cell type. respiratory
- Cell-matrix signals (integrin β1) specify bone endothelial cells during developmental osteogenesis[6] “Blood vessels in the mammalian skeletal system control bone formation and support haematopoiesis by generating local niche environments. Here, we report that embryonic and early postnatal long bone contains a specialized endothelial cell subtype, termed type E, which strongly supports osteoblast lineage cells and later gives rise to other endothelial cell subpopulations. The differentiation and functional properties of bone endothelial cells require cell-matrix signalling interactions." Bone Development | INTEGRIN BETA-1
- foxl3 is a germ cell-intrinsic factor involved in sperm-egg fate decision in medaka[7] "Sex determination is an essential step in the commitment of a germ cell to a sperm or egg. However, the intrinsic factors that determine the sexual fate of vertebrate germ cells are unknown. Here we show that foxl3, which is expressed in germ cells but not somatic cells in the gonad, is involved in sperm-egg fate decision in medaka fish. Adult XX medaka with disrupted foxl3 developed functional sperm in the expanded germinal epithelium of a histologically functional ovary. In chimeric medaka, mutant germ cells initiated spermatogenesis in female wild-type gonad. These results indicate that a germ cell-intrinsic cue for the sperm-egg fate decision is present in medaka and that spermatogenesis can proceed in a female gonadal environment." ( Forkhead Transcription Factor foxl3 is an ancient duplicated copy of foxl2 | Medaka Development | Oocyte Development | Spermatozoa Development)
|
| Older papers
|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
- The Encyclopedia of DNA Elements (ENCODE) project Wed, Sep 5, 2012 "The Human Genome Project produced an almost complete order of the 3 billion pairs of chemical letters in the DNA that embodies the human genetic code — but little about the way this blueprint works. Now, after a multi-year concerted effort by more than 440 researchers in 32 labs around the world, a more dynamic picture gives the first holistic view of how the human genome actually does its job." NIH Nature
- Properties of developmental gene regulatory networks[8] Introductory paper from the "Gene Networks in Development and Evolution Special Feature Sackler Colloquium" (2008) "The modular components, or subcircuits, of developmental gene regulatory networks (GRNs) execute specific developmental functions, such as the specification of cell identity. We survey examples of such subcircuits and relate their structures to corresponding developmental functions. These relations transcend organisms and genes, as illustrated by the similar structures of the subcircuits controlling the specification of the mesectoderm in the Drosophila embryo and the endomesoderm in the sea urchin, even though the respective subcircuits are composed of nonorthologous regulatory genes."
|
Movies
Starting Out
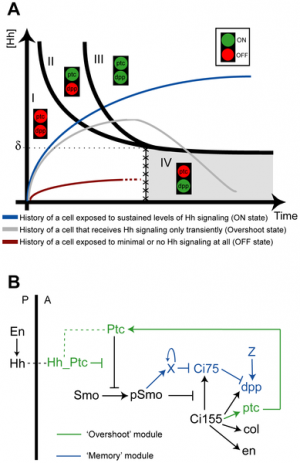
Model of Sonic hedgehog signaling in the drosophila wing
[9]There are several different ways to begin to look at molecular development.
- look at the earliest molecular events involved in patterning the differentiation of cells- axis formation.
- or look at early events in patterning with differentiation and migration of cells to form the trilaminar embryo (gastrulation).
- or look at the signaling mechanisms and the different types of factors involved.
- or look at their role in the development of specific systems.
Signaling during development, though complex, can also be grouped into a few specific classes. These mechanisms have also been listed and described briefly on Signaling Mechanisms page.
For this section, Molecular Biology of the Cell (now indexed and available online from PubMed) and the extracted Cell Biology version are good reference texts. Also the key journals in this area are: Development, Genes and Development, PNAS, EMBOJ.
There is also detailed molecular information available from the abnormalities pages where there are links to the OMIM Database entry for specific genetic disorders.
Gene Expression
DNA -> RNA -> Protein
Our genetic information is stored within each cell in cell organelles of the nucleus and mitochondria.
- In the nucleus as chromosomes.
- In the mitochondria as circular DNA.
Gene expression can often simply be described by the pathway: DNA -> mRNA -> Protein. While each cell contains the same genetic material, not all genes are ever expressed and the subset of genes expressed at any one time affects cellular differentiation and mature cell function. This can be impacted by both signaling and epigenetic mechanisms.
The human chromosomes consist of proteins and nucleic acids, the common nucleic acids or bases that forms DNA can be represented by a standardised IUPAC single letter code.
Deoxyribonucleic acid
Deoxyribonucleic acid (DNA) consists of 4 nucleotides (bases):
- Adenine (A)
- Cytosine (C)
- Guanine (G)
- Thymine (T)
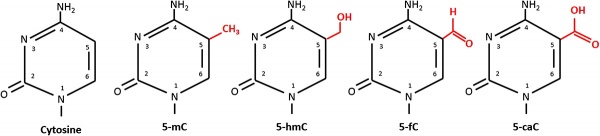
An additional amino acid, 5-Formylcytosine (5fC), a rare nucleotide has recently been identified as a stable DNA modification occurring in mammals.[10] The function of this additional base was thought to be involved only in active DNA demethylation, but may also have roles in gene regulation.
Ribonucleic acid
Ribonucleic acid (RNA) consists of 4 nucleotides:
- Adenine (A)
- Cytosine (C)
- Guanine (G)
- Uracil (U)
The original "RNA family" consisted of just 3 main members; transfer RNA (tRNA), ribosomal RNA (rRNA) and messenger RNAs (mRNA). Involved in gene expression through protein translation (synthesis).
In recent years this RNA family, shown in the table below, has been expanded to include newly identified members: small nuclear RNA (snRNA) , small nucleolar RNA (snoRNA), and short regulatory RNA (piwi-associated RNA (piRNA), endogenous short-interfering RNA (endo-siRNA) and microRNA (miRNA) and now long non-coding RNA (lncRNA). Each of these new family members has a range of potential roles in development and differentiation.
| RNA Class |
Acronym |
Roles |
Examples |
Reviews
|
| transfer RNA |
tRNA |
carry amino acid in cell cytoplasm to ribosome |
|
PMID 24966867 PMID 27095039
|
| messenger RNA |
mRNA |
transcribed from DNA in cell nucleus and relocate to cytoplasm for translation on the ribosome |
Example |
PMID 28102265
|
| ribosomal RNA |
rRNA |
structural RNA that allow the assembly of ribosomal proteins in the cytoplasm into ribosomal subunits required for protein translation |
Example |
PMID 30356013
|
| small nuclear RNA |
snRNA |
involved in cell nucleus RNA splicing |
|
PMID 23980890
|
| small nucleolar RNA |
snoRNA |
modify other small RNAs (rRNAs and tRNAs) |
box C/D snoRNAs and the box H/ACA |
PMID 21664409 PMID 19446021
|
| microRNA |
miRNA |
post-transcriptional regulator of gene expression |
|
PMID 25128264
|
| endogenous short-interfering RNA |
endo-siRNA |
short regulatory RNA |
mainly characterised in plants |
PMID 22578318
|
| piwi-associated RNA |
piRNA |
short regulatory RNA |
|
PMID 18032451 PMID 22103557
|
| long non-coding RNA |
ncRNA |
non-coding RNA greater than 200bp in length may have different roles in signalling, protein processing and differentiation |
|
PMID 24829860
|
- Links: Molecular Development - Ribonucleic acid | microRNA
Protein
The individual amino acids that form all proteins can be represented by a standardised single letter code, or three letter code or by their entire name.
Single Letter Code
A - Alanine (Ala) | C - Cysteine (Cys) | D - Aspartic Acid (Asp) | E - Glutamic Acid (Glu) | F - Phenylalanine (Phe) | G - Glycine (Gly) | H - Histidine (His) | I - Isoleucine (Ile) | K - Lysine (Lys) |L - Leucine (Leu) | M - Methionine (Met) | N - Asparagine (Asn) | P - Proline (Pro) | Q - Glutamine (Gln) | R - Arginine (Arg) | S - Serine (Ser) | T - Threonine (Thr) | V - Valine (Val) | W - Tryptophan (Trp) | Y - Tyrosine (Tyr)
Hydropathy Index
- a number representing the hydrophobic or hydrophilic properties of the amino acid sidechain.
- larger the number the more hydrophobic the amino acid.
- most hydrophobic amino acids are isoleucine (4.5) and valine (4.2), hydrophobic amino acids tend to be internal
- most hydrophilic amino acids are arginine (-4.5) and lysine (-3.9), hydrophilic amino acids are more commonly found towards the protein surface.
- Links: Protein
Signaling Factors
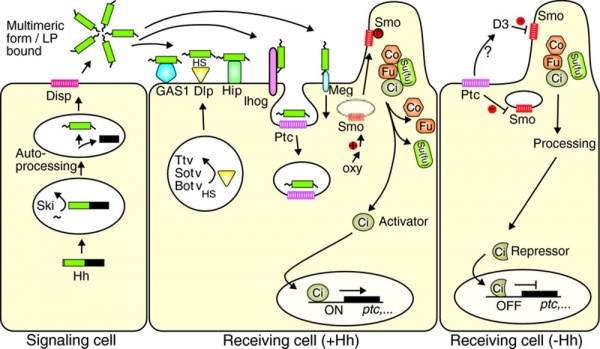
Hedgehog signaling pathway[12]
Note spelling difference: USA signaling, UK signalling.
Axis Formation
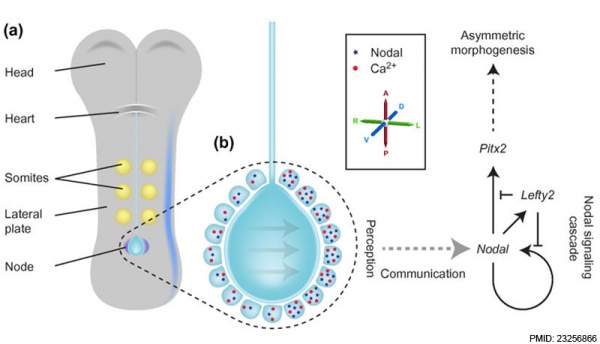
Embryo left-right asymmetry pathway[13]
The mechanisms that specify location within the embryo, therefore establishing axes, appears to be controlled by similar signaling mechanisms in different species.
- Links: Axis Formation
Transcription Factors
Transcription factors act by activating or inhibiting transcription either a single gene or more commonly acting on a family of genes. The transcription factor binding sites can be upstream of affected genes or some distance away from the initiation codon and many of these individual factors belong to families of many factors that share common DNA binding motifs.
- forkhead box (FOX) superfamily contain a DNA-binding motif known as the forkhead box or winged-helix domain.
Epigenetics
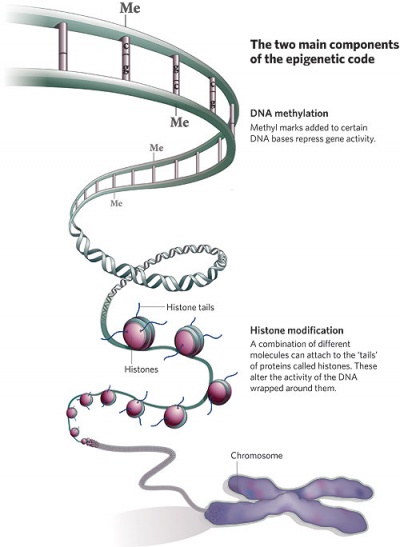
| Epigenetics as the name implies, is the inheritance mechanisms that lie outside the DNA sequence of our genes.
One of the initial discoveries was the effects of DNA methylation upon gene expression and then modifications of nucleosomal histones.
This DNA methylation, usually associated with 5-methylcytosine (m5C), leads to transcriptional silencing in vertebrates.
- Links: Epigenetics
|
|
References
- ↑ Qiu J. (2006). Epigenetics: unfinished symphony. Nature , 441, 143-5. PMID: 16688142 DOI.
- ↑ 2.0 2.1 Turner FR & Mahowald AP. (1979). Scanning electron microscopy of Drosophila melanogaster embryogenesis. III. Formation of the head and caudal segments. Dev. Biol. , 68, 96-109. PMID: 108157
- ↑ Lammers NC, Galstyan V, Reimer A, Medin SA, Wiggins CH & Garcia HG. (2020). Multimodal transcriptional control of pattern formation in embryonic development. Proc. Natl. Acad. Sci. U.S.A. , 117, 836-847. PMID: 31882445 DOI.
- ↑ Chen X, Ke Y, Wu K, Zhao H, Sun Y, Gao L, Liu Z, Zhang J, Tao W, Hou Z, Liu H, Liu J & Chen ZJ. (2019). Key role for CTCF in establishing chromatin structure in human embryos. Nature , , . PMID: 31801998 DOI.
- ↑ Little DR, Gerner-Mauro KN, Flodby P, Crandall ED, Borok Z, Akiyama H, Kimura S, Ostrin EJ & Chen J. (2019). Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2-1. Proc. Natl. Acad. Sci. U.S.A. , 116, 20545-20555. PMID: 31548395 DOI.
- ↑ Langen UH, Pitulescu ME, Kim JM, Enriquez-Gasca R, Sivaraj KK, Kusumbe AP, Singh A, Di Russo J, Bixel MG, Zhou B, Sorokin L, Vaquerizas JM & Adams RH. (2017). Cell-matrix signals specify bone endothelial cells during developmental osteogenesis. Nat. Cell Biol. , 19, 189-201. PMID: 28218908 DOI.
- ↑ Nishimura T, Sato T, Yamamoto Y, Watakabe I, Ohkawa Y, Suyama M, Kobayashi S & Tanaka M. (2015). Sex determination. foxl3 is a germ cell-intrinsic factor involved in sperm-egg fate decision in medaka. Science , 349, 328-31. PMID: 26067255 DOI.
- ↑ Davidson EH & Levine MS. (2008). Properties of developmental gene regulatory networks. Proc. Natl. Acad. Sci. U.S.A. , 105, 20063-6. PMID: 19104053 DOI.
- ↑ Nahmad M & Stathopoulos A. (2009). Dynamic interpretation of hedgehog signaling in the Drosophila wing disc. PLoS Biol. , 7, e1000202. PMID: 19787036 DOI.
- ↑ Martin Bachman, Santiago Uribe-Lewis, Xiaoping Yang, Heather E Burgess, Mario Iurlaro, Wolf Reik, Adele Murrell and Shankar Balasubramanian. 5-Formylcytosine can be a stable DNA modification in mammals Nature Chemical Biology (2015) doi:10.1038/nchembio.1848.
- ↑ <pubmed>7108955</pubmed>
- ↑ Bürglin TR. (2008). The Hedgehog protein family. Genome Biol. , 9, 241. PMID: 19040769 DOI.
- ↑ Norris DP. (2012). Cilia, calcium and the basis of left-right asymmetry. BMC Biol. , 10, 102. PMID: 23256866 DOI.
Terms
| Genetic Terms (expand to view)
|
genetic abnormalities | Molecular Development | meiosis | mitosis
- Alpha-Fetoprotein test (APF test) A prenatal test to measure the amount of a fetal protein in the mother's blood (or amniotic fluid). Abnormal amounts of the protein may indicate genetic or developmental problems in the fetus. Serum alpha-fetoprotein (AFP) is a fetal glycoprotein produced by the yolk sac and fetal liver. Low levels of AFP normally occur in the blood of a pregnant woman, high levels may indicate neural tube defects (spina bifida, anencephaly). (More? Alpha-Fetoprotein)
- anaphase B - Cell division term referring to the part of anaphase during which the poles of the mitotic spindle move apart. (More? mitosis)
- antisense - a sequence of DNA that is complementary usually to coding sequence of DNA or mRNA. Has been used experimentally to perturb or block gene expression. Also a mechanism that has been found to occur naturally as a regulatory mechanism.
- aneuploidy - Genetic term used to describe an abnormal number of chromosomes mainly (90%) due to chromosome malsegregation mechanisms in maternal meiosis I.
- autosomal inheritance - some hereditary diseases are described as autosomal which means that the disease is due to a DNA error in one of the 22 pairs that are not sex chromosomes. Both boys and girls can then inherit this error. If the error is in a sex chromosome, the inheritance is said to be sex-linked.
- base - another term for nucleotide (usually a t c g).
- base pair - Double stranded DNA has nucleotides A-T, C-G, paired by hydrogen bonds (2 for AT, 3 for GC). Note this means that GC is harder to separate that AT.
- cis-acting elements - DNA sequences that through transcription factors or other trans-acting elements or factors, regulate the expression of genes on the same chromosome.
- cohesin - a multi-protein subunit complex required to keep the sister chromatids together until their separation at anaphase (both in mitosis and meiosis), can also form rings that connect two DNA segments.
- copy number variation - (copy number variants,CNVs) a DNA segment of one kilobase (kb) or larger that is present at a variable copy number in comparison with a reference genome. (More? Nature)
- disomy - Genetic term referring to the presence of two chromosomes of a homologous pair in a cell, as in diploid. See chromosomal number genetic disorders uniparental disomy and aneuploidy. Humans have pairs usually formed by one chromosome from each parent.
- DNMT - DNA methyltransferase.
- DNA - DeoxyriboNucleic Acid. The genetic material found in mammalian chromosomes and mitochondria. Consisting of 4 nucleic acids (ATCG) that combine in a triptych (3 nucleotide codon) code for protein amino acids (3nt = 1aa).
- DNA duplex - double stranded base-paired DNA forming a helix.
- dominant inheritance - With autosomal dominant inheritance, there is an error in one of the 22 chromosome pairs. But the damaged gene dominates over the normal gene received from the other parent. If one of the parents has a disease caused by an autosomal dominant gene, all the children will have a 50 per cent risk of inheriting the dominant gene and a 50 per cent chance of not inheriting it. The children who do not inherit the damaged dominant gene will not themselves suffer from the disease, nor will they be able to pass the gene on to future children. This type of inheritance is present for example in Huntington's disease.
- Down Syndrome - The historic name used for Trisomy 21, named after the original identifier Down, J.L.H. in a 1866 paper (cited above).
- enhancer - A cis-regulatory sequence that can regulate levels of transcription from an adjacent promoter. Many tissue-specific enhancers can determine spatial patterns of gene expression in higher eukaryotes. Enhancers can act on promoters over many tens of kilobases of DNA and can be 5' or 3' to the promoter they regulate.
- exon - a block of protein encoding sequence of DNA in a gene. Many proteins are made of several exons "stitched" or spliced together by editing out non-coding (intron) sequences.
- fasta - a format for listing DNA sequence, where the first line has descritive information followed on the next line by the sequence without numbering.
- GC repeat - a string of GC sequence repeated several times. Also associated with GC expansion, a mutational process that may lead eventually to serious gene expression effects.
- gene - a sequence of DNA that encodes an individual protein.
- genetic code - the 3 nucleotide sequence that forms a codon for a single amino acid or stop. See the gene code.
- genome - the complete genetic information in the form of DNA available to a specific species.
- hairpin loop - a folding of RNA generated by base pairing making a "===()" structure, the end loop and or stem of this structure can then interact with proteins or other RNA.
- heteroplasmy - within a cell when more than one type of mitochondrial DNA (mtDNA) genome exists within the mitochondrion, or between mitochondria. Heteroplasmy can generate a pathogenic mutation, for example transfer RNA leucine (tRNALeu(UUR)) 3243A > G mutant can result in diabetes.
- HyperD - hypermethylated domain has a role in regulating gene expression and epigenomics. The early embryo has large alterations in methylation patterns and DNA modification. (More? PMID 1943996)
- HypoD - hypomethylated domain has a role in regulating gene expression and epigenomics. The early embryo has large alterations in methylation patterns and DNA modification. (More? PMID 1943996)
- igDMR - imprinted germline differentially methylated regions
- intron - a block of DNA within a gene not encoding a protein. Edited, spliced, out during transcription into mRNA. Originally thought not to contain any information, but more and more this appears not to be the case. Some intron sequences have been shown to regulate gene expression during development (eg c elegans, Lin 14)
- karyotype - [Greek, karyon = kernel or nucleus + typos= stamp] The chromosomal makeup of a cell. Karyotyping describes the clinical genetic test.
- meiosis I (MI) The first part of meiosis resulting in separation of homologous chromosomes, in humans producing two haploid cells (N chromosomes, 23), a reductional division.
- Meiosis I: Prophase I - Metaphase I - Anaphase I - Telophase I
- meiosis II - (MII) The second part of meiosis. In male human spermatogenesis, producing of four haploid cells (23 chromosomes, 1N) from the two haploid cells (23 chromosomes, 1N), each of the chromosomes consisting of two sister chromatids produced in meiosis I. In female human oogenesis, only a single haploid cell (23 chromosomes, 1N) is produced. Meiosis II: Prophase II - Metaphase II - Anaphase II - Telophase II
- mosaic autosome monosomy - genetic abnormality caused by embryonic fusion or loss of an autosome early in embryonic development, resulting in a subset of cells in the body having only one of a pair of autosomes. ICD-11 LD43.1 Mosaic monosomy of autosome
- mRNA - messenger, transcribed from DNA in the nucleus and in mitochondria. Is translated by the ribosome in the cytoplasm (or mitochondrial matrix). Intermediate step in gene expression. (DNA-> mRNA-> protein).
- mutation - any process which results in the alteration of the DNA sequence. Some conservative mutations may have no effect on the final amino acid encoded.
- nuchal translucency - (fetal nuchal-translucency thickness) An initial Trisomy 21 diagnostic ultrasound measurement in the fetal neck region carried out by trans-abdominal ultrasound at gestational age GA 10–14 weeks. Fetal sagittal section scan at a magnification that the fetus occupied at least 75% of the image. Measured is the maximum thickness of the subcutaneous translucency between the skin and the soft tissue overlying the cervical spine.
- Philadelphia chromosome - (Philadelphia translocation) Genetic term referring to a chromosomal abnormality resulting from a reciprocal translocation between chromosome 9 and 22 (t(9;22)(q34;q11)). This is associated with the disease chronic myelogenous leukemia (CML).
- ploidy - refers to the chromosomal genetic content of cells.
- promoter - A regulatory region a short distance upstream from the 5' end of a transcription start site that acts as the binding site for RNA polymerase II. A region of DNA to which RNA polymerase IIbinds in order to initiate transcription.
- point mutation - a change in a single nucleotide.
- recessive inheritance - With autosomal recessive inheritance, the diseased individual has inherited the same gene damage from both father and mother. The damage is found on both chromosomes in the pair. But as this is not ´dominant gene damageª, neither father nor mother show any sign of disease, they are healthy carriers of the gene. We are all carriers of about five recessive genes of this type, but as spouses are seldom carriers of exactly the same damaged gene(s), all will probably go well in the next generation.
- regulatory sequence - (regulatory region, regulatory area) is a segment of DNA where regulatory proteins such as transcription factors bind preferentially.
- ribosome - complex of rRNA and ribosomal proteins, bind mRNA and translate it into protein.
- RNA - RiboNucleic Acid. The intermediate nucleic acid involved in gene expression. It comes in 3 forms: tRNA, mRNA, rRNA.
- rRNA - ribosomal, translates mRNA into protein. rRNA provides the "scaffolding" on which many ribosomal proteins are assembled as 2 subunits that themselves assemble to form a ribosome. rRNA genes are localized to the nucleolus in the nucleus, a sometimes visible region of DNA usually constantly being transcribed.
- segmental aneuploidies - generated when a small piece of a chromosome is gained or lost during cell division, resulting in subchromosomal copy number (CN) changes.
- single umbilical artery - (SUA) Placental cord with only a single placental artery (normally paired). This abnormality can be detected by ultrasound (colour flow imaging of the fetal pelvis) and is used as an indicator for further prenatal diagnostic testing for chromosomal abnormalities and other systemic defects. (More? Prenatal Diagnosis | Ultrasound)
- telophase - Cell division term referring to the fifth mitotic stage, where the vesicles of the nuclear envelope reform around the daughter cells, the nucleoli reappear and the chromosomes unfold to allow gene expression to begin. This phase overlaps with cytokinesis, the division of the cell cytoplasm.
- telomere - regions at the end of chromosomes. Shortening of the telomeres is thought to be associated with cellular aging. The enzyme that maintains the telomere is called telomerase. Introducing this gene into a cell can extend the cells lifespan.
- topologically associating domain - (TAD) a self-interacting genomic region, DNA sequences within a TAD physically interact with each other more frequently than with sequences outside the TAD.
- transcription factor - a protein which binds to DNA activating (usually) gene expression. There are many different ways and forms that this activation can take place, but most transcription factors fall into specific classes (eg zinc fingers, helix loop helix).
- triple markers - alpha-fetoprotein, human chorionic gonadotropin, and unconjugated estriol. ))trisomy 21}}
- triploidy - genetic abnormality caused by one additional set of chromosomes, for a total of 69 chromosomes. Maternally present with albuminuria, edema, or hypertension. Extra maternally inherited chromosomes, microcephaly and an enlarged placenta that is enlarged and filled with cysts. Extra paternally inherited chromosomes, severe growth problems, enlarged head, and a small placenta. Non-mosaic triploidy is highly lethal, and is rarely observed in live births. ICD-11 LD42.0 Triploidy
- trisomy mosaicism - a rare chromosome disorder characterized by having an extra copy of a chromosome in a proportion, but not all, of a person’s cells.
- tRNA - transfer, binds single amino acids acts as a "donor' for protein synthesis.
- uniparental disomy - Genetic term referring to cells containing both copies of a homologous pair of chromosomes from one parent and none from the other parent.
|
|
|
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
Gene Ontology Consortium
- Gene Ontology Consortium - "The mission of the GO Consortium is to develop a comprehensive, computational model of biological systems, ranging from the molecular to the organism level, across the multiplicity of species in the tree of life."
- NHGR - The ENCODE Project ENCyclopedia Of DNA Elements
- Foldit - Amino Acids
- Cytoscape - open source software platform for visualizing complex networks and integrating these with any type of attribute data.
- iRegulon - Detects master regulators and cis-regulatory interactions.
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 28) Embryology Molecular Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Molecular_Development
- What Links Here?
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G









