Molecular Development - microRNA
| Embryology - 27 Feb 2026 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
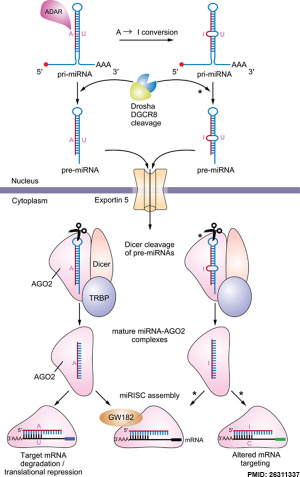
Micro RNA (miRNA) are 22 nucleotide non-coding RNAs that regulate about 30% of mammalian genes.
The original "RNA family" consisted of just 3 main members; transfer RNA (tRNA), ribosomal RNA (rRNA) and messenger RNAs (mRNA).
These small pieces of RNA have been identified as important negative regulators in both development and adult cell processes involving gene expression. The repressive effects are typically relatively mild, with parallel regulatory mechanisms, the phenotype may require a stress condition or tissue injury for their expression.
miRNAs are initially transcribed by RNA polymerase II as part of capped and polyadenylated primary transcripts (pri-miRNAs) that can be either protein-coding or non-coding. The primary transcript is cleaved by the Drosha ribonuclease III enzyme to produce an approximately 70-nt stem-loop precursor miRNA (pre-miRNA), which is further cleaved by the cytoplasmic Dicer ribonuclease to generate the mature ~22 (20-24 nt) nucleotide miRNA and antisense miRNA star (miRNA*) products.
The miRNAs then bind (by imperfectly complementation) usually to 3′ untranslated region of target mRNAs, resulting in mRNA degradation and/or translational inhibition.
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: microRNA Embryology <pubmed limit=5>microRNA Embryology</pubmed> |

Pathway
- Primary miRNA precursors are transcribed in the nucleus
- Processed into 19-22 nt mature miRNAs by
- nuclear RNase III enzyme Drosha and its co-factor Dgcr8
- cytoplasmic RNase III enzyme Dicer
- miRNAs then loaded into the miRNA-induced silencing complex (miRISC)
Some miRNA subclasses are either Drosha- or Dicer-independent.
miRNA-Induced Silencing Complex
(miRISC) components include the argonaute proteins (Ago1–4), Ago2 is the only protein with the slicer activity.
Functional miRNA Grouping
The following identifies microRNAs that have been identified with specific developmental processes, based upon a commercial collation of basic research data.
- Pluripotency
- let-7a, let-7b, let-7c, let-7d, let-7e, let-7g, miR-101, miR-106b, miR-125b, miR-130a, miR-133b, miR-141, miR-15a, miR-17, miR-182, miR-183, miR-18a, miR-18b, miR-205, miR-20a, miR-20b, miR-21, miR-214, miR-22, miR-222, miR-23b, miR-24, miR-302a, miR-302c, miR-345, miR-424, miR-498, miR-518b, miR-520g.
- miR-10 - microRNA precursor part of an RNA gene family which contains miR-10, miR-51, miR-57, miR-99 and miR-100. miR-10, miR-99 and miR-100 have been confirmed in a wide range of species.
Nomenclature
arm the mature sequence
- left (-5p)
- right (-3p)
Early Development
- Embryoid Bodies
- miR-132, miR-181a, miR-9.
- Definitive Endoderm
- miR-205, miR-375.
Ectoderm
- Neural Development
- let-7b, miR-103a, miR-106b, miR-10b, miR-124, miR-125b, miR-130a, miR-132, miR-134, miR-137, miR-16, miR-181a, miR-182, miR-183, miR-20a, miR-210, miR-219-5p, miR-22, miR-23b, miR-24, miR-26a, miR-302a, miR-302c, miR-7, miR-9, miR-96.
- Neural Tube - miRNA-430 organises cell division (planes) for neural tube morphogenesis. PMID 26658217
- Eye Development
- miR-130a, miR-196a, miR-219-5p, miR-23b, miR-96.
- Epidermal Differentiation
- let-7b, miR-205, miR-210, miR-23b, miR-26a.
- Inner Ear Development
- miR-182, miR-183, miR-96.
- Integumentary Development
- 70 miRNAs are expressed in embryonic skin.
- miR-205 in epithelia and promotes keratinocyte migration in vitro
- miR-203 in suprabasal epidermal cells
Mesoderm
- Haematopoiesis
- let-7e, miR-125a-5p, miR-142-3p, miR-223.
- T Cell Development
- let-7a, let-7f, miR-106b, miR-142-5p, miR-146b-5p, miR-150, miR-15a, miR-15b, miR-16, miR-181a, miR-20a, miR-222, miR-26a.
- Erythropoiesis
- let-7a, let-7b, let-7c, let-7d, let-7f, let-7g, let-7i, miR-126, miR-128a, miR-137, miR-155, miR-15b, miR-16, miR-17, miR-181a, miR-182, miR-185, miR-206, miR-21, miR-22, miR-222, miR-24, miR-26a, miR-96.
- Lymphopoiesis
- let-7b, miR-125b, miR-128a, miR-16, miR-181a, miR-21, miR-24.
- Megakaryopoiesis
- miR-106b, miR-10a, miR-10b, miR-122, miR-126, miR-127-5p, miR-129-5p, miR-134, miR-146a, miR-150, miR-155, miR-17, miR-18a, miR-192, miR-20a, miR-20b, miR-21, miR-22, miR-301a, miR-33a, miR-378, miR-92a, miR-93.
- Monocyte Differentiation
- miR-155, miR-222, miR-424.
- Myelopoiesis
- miR-103a, miR-128a, miR-17, miR-181a, miR-24.
- Angiogenesis
- miR-126, miR-130a, miR-218, miR-222, miR-92a.
- Myogenesis
- miR-1, miR-125b, miR-206, miR-26a.
- Osteogenesis
- miR-141, miR-15b, miR-424.
- Adipogenesis
- let-7b, let-7c, let-7e, miR-100, miR-101, miR-103a, miR-10b, miR-146b-5p, miR-155, miR-182, miR-192, miR-194, miR-196a, miR-21, miR-210, miR-214, miR-22, miR-24, miR-498, miR-96, miR-99a.
- Chondrogenesis: let-7f, miR-1, miR-132, miR-181a, miR-196a, miR-96, miR-99a.
- Heart Development: miR-1, miR-208, miR-488.
Endoderm
- Liver Development
- let-7a, let-7b, let-7c, miR-10a, miR-122, miR-125b, miR-192, miR-21, miR-22, miR-23b, miR-92a, miR-99a.
- Pancreatic Development
- miR-15a, miR-15b, miR-16, miR-195, miR-214, miR-375, miR-7, miR-9.
- Intestinal Development
- let-7d, let-7e, miR-103a, miR-106b, miR-125b, miR-126, miR-130a, miR-141, miR-146b-5p, miR-17, miR-192, miR-194, miR-21, miR-215, miR-301a, miR-424.
- Data: SABiosciences Cell Differentiation & Development miRNA PCR Array
Nucleolus
Five miRNAs show highest nucleolar concentration in myoblasts using the microarray assay, miR-340-5p, miR-351, miR-494, miR-664, or let-7e, are thought to be skeletal muscle-specific miRNAs (i.e., miR-1, miR-133, and miR-206 and perhaps miR-95, miR-128a, and miR-499).[9]
Sex Determination
microRNA shown in mouse genital development (miR-199-214, miR-182-183-96, and miR-34c-5p)[5]
Hearing
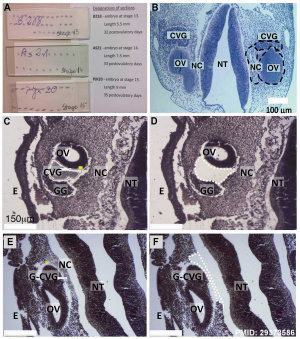
- miR-183 family (miR-96, miR-182, and miR-183) are differentially expressed in the cochleovestibular ganglion compared to neural crest and otic vesicle at Carnegie stage 13. From a recent study of microRNAs in the human embryonic inner ear during Carnegie developmental stages 13, 14 and 15.[2]
- miR-194 regulates the development and differentiation of sensory patches and statoacoustic ganglion of inner ear by fgf4.
From an earlier study in mouse[10] and a recent study of microRNAs in the zebrafish embryonic inner ear.[11]
Other RNAs
The original "RNA family" consisted of just 3 main members; transfer RNA (tRNA), ribosomal RNA (rRNA) and messenger RNAs (mRNA). MicroRNA (miRNA) is a member of other newly identified classes of RNA have been suggested to have many different roles in signalling, protein processing and differentiation.
- long non-coding RNA - (lncRNA) A class of RNA greater than 200bp in length that do not encode a protein product.[12]
- small nuclear RNA - (snRNA)
- small nucleolar RNA - (snoRNA)
- short regulatory RNA (piwi-associated RNA (piRNA)
- endogenous short-interfering RNA - (endo-siRNA)
- microRNA - (miRNA)
References
- ↑ 1.0 1.1 Deiuliis JA. (2016). MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. Int J Obes (Lond) , 40, 88-101. PMID: 26311337 DOI.
- ↑ 2.0 2.1 2.2 Chadly DM, Best J, Ran C, Bruska M, Woźniak W, Kempisty B, Schwartz M, LaFleur B, Kerns BJ, Kessler JA & Matsuoka AJ. (2018). Developmental profiling of microRNAs in the human embryonic inner ear. PLoS ONE , 13, e0191452. PMID: 29373586 DOI.
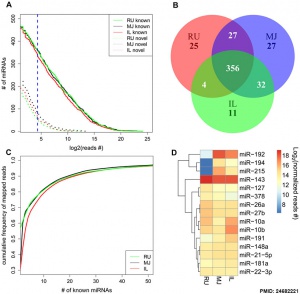
- ↑ 3.0 3.1 3.2 Liang G, Malmuthuge N, McFadden TB, Bao H, Griebel PJ, Stothard P & Guan le L. (2014). Potential regulatory role of microRNAs in the development of bovine gastrointestinal tract during early life. PLoS ONE , 9, e92592. PMID: 24682221 DOI.
- ↑ Svoboda P. (2017). Long and small noncoding RNAs during oocyte-to-embryo transition in mammals. Biochem. Soc. Trans. , 45, 1117-1124. PMID: 28939692 DOI.
- ↑ 5.0 5.1 Fernández-Pérez D, Brieño-Enríquez MA, Isoler-Alcaraz J, Larriba E & Del Mazo J. (2018). MicroRNA dynamics at the onset of primordial germ and somatic cell sex differentiation during mouse embryonic gonad development. RNA , 24, 287-303. PMID: 29187591 DOI.
- ↑ Reliszko ZP, Gajewski Z & Kaczmarek MM. (2017). Signs of embryo-maternal communication: miRNAs in the maternal serum of pregnant pigs. Reproduction , 154, 117-128. PMID: 28592665 DOI.
- ↑ Gilchrist GC, Tscherner A, Nalpathamkalam T, Merico D & LaMarre J. (2016). MicroRNA Expression during Bovine Oocyte Maturation and Fertilization. Int J Mol Sci , 17, 396. PMID: 26999121 DOI.
- ↑ Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S & Diamond MP. (2011). A survey of small RNAs in human sperm. Hum. Reprod. , 26, 3401-12. PMID: 21989093 DOI.
- ↑ Politz JC, Hogan EM & Pederson T. (2009). MicroRNAs with a nucleolar location. RNA , 15, 1705-15. PMID: 19628621 DOI.
- ↑ Wang XR, Zhang XM, Zhen J, Zhang PX, Xu G & Jiang H. (2010). MicroRNA expression in the embryonic mouse inner ear. Neuroreport , 21, 611-7. PMID: 20467336 DOI.
- ↑ Cao H, Shi J, Du J, Chen K, Dong C, Jiang D & Jiang H. (2018). MicroRNA-194 Regulates the Development and Differentiation of Sensory Patches and Statoacoustic Ganglion of Inner Ear by Fgf4. Med. Sci. Monit. , 24, 1712-1723. PMID: 29570699
- ↑ Goff LA & Rinn JL. (2015). Linking RNA biology to lncRNAs. Genome Res. , 25, 1456-65. PMID: 26430155 DOI.
Reviews
Wan Y, Kertesz M, Spitale RC, Segal E & Chang HY. (2011). Understanding the transcriptome through RNA structure. Nat. Rev. Genet. , 12, 641-55. PMID: 21850044 DOI.
Sayed D & Abdellatif M. (2011). MicroRNAs in development and disease. Physiol. Rev. , 91, 827-87. PMID: 21742789 DOI.
Yi R & Fuchs E. (2011). MicroRNAs and their roles in mammalian stem cells. J. Cell. Sci. , 124, 1775-83. PMID: 21576351 DOI.
Agirre E & Eyras E. (2011). Databases and resources for human small non-coding RNAs. Hum. Genomics , 5, 192-9. PMID: 21504869
Suh N & Blelloch R. (2011). Small RNAs in early mammalian development: from gametes to gastrulation. Development , 138, 1653-61. PMID: 21486922 DOI.
Ghildiyal M & Zamore PD. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. , 10, 94-108. PMID: 19148191 DOI.
Articles
Mujahid S, Logvinenko T, Volpe MV & Nielsen HC. (2013). miRNA regulated pathways in late stage murine lung development. BMC Dev. Biol. , 13, 13. PMID: 23617334 DOI.
Dong J, Jiang G, Asmann YW, Tomaszek S, Jen J, Kislinger T & Wigle DA. (2010). MicroRNA networks in mouse lung organogenesis. PLoS ONE , 5, e10854. PMID: 20520778 DOI.
Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH & Huang HD. (2008). miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. , 36, D165-9. PMID: 18029362 DOI.
Search Pubmed
Search Pubmed Now: microRNA
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- miRBase - http://www.mirbase.org
- miRIAD - http://www.bioinfo.mochsl.org.br/miriad/miRNA/hsa-mir-1180/ is a web search tool designed to help users to access integrated information concerning intragenic microRNAs and their host genes
- Memorial Sloan-Kettering Cancer Center - microRNA.org - Targets and Expression
- SABiosciences - Cell Differentiation & Development miRNA PCR Array
- Sigma-Aldrich - miRNA Introduction
- miRNAMap - http://mirnamap.mbc.nctu.edu.tw/
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2026, February 27) Embryology Molecular Development - microRNA. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Molecular_Development_-_microRNA
- © Dr Mark Hill 2026, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G
