Developmental Signals - Notch
| Embryology - 28 Feb 2026 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction

The notch proteins were first identified in drosophila development and have since been identified as regulators of cell fate decisions during development. These are a family of cell surface transmembrane receptors that pass once through the plasma membrane.
This signalling pathway is involved with many different developmental patterning pathways and in the adult is a key regulator in the vascular system.
- Notch Links: Notch structure cartoon | Notch signaling pathway cartoon | Notch and signaling pathway cartoon | Developmental Signals - Notch | Molecular Factors
| Factor Links: AMH | hCG | BMP | sonic hedgehog | bHLH | HOX | FGF | FOX | Hippo | LIM | Nanog | NGF | Nodal | Notch | PAX | retinoic acid | SIX | Slit2/Robo1 | SOX | TBX | TGF-beta | VEGF | WNT | Category:Molecular |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Notch |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Notch Signaling
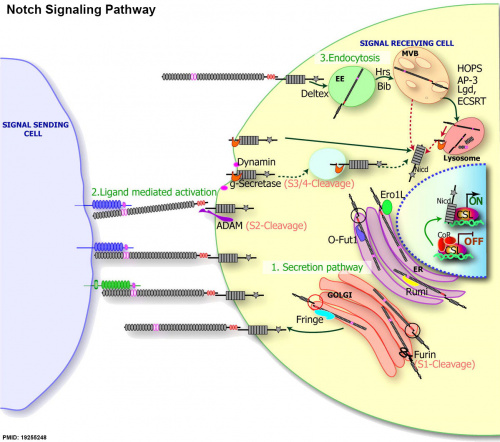
|
| Notch signaling pathway[1] |
(text from original figure legend) |
Notch Receptors
Notch receptors belong to the Ankyrin repeat domain containing (ANKRD) gene family (242 members).
| Table - Human Notch Family (Ankyrin repeat domain - ANKRD) | |||||
| Approved Symbol |
Approved Name | Previous Symbols |
Synonyms | Chromosome | OMIM ID |
|---|---|---|---|---|---|
| NOTCH1 | notch 1 | TAN1 | 9q34.3 | 190198 | |
| NOTCH2 | notch 2 | 1p12 | 600275 | ||
| NOTCH3 | notch 3 | CADASIL | CASIL | 19p13.12 | 600276 |
| NOTCH4 | notch 4 | INT3 | 6p21.32 | 164951 | |
| Links: Notch | OMIM BMP2 | HGNC - ANKRD | Bmp Family | Notch Family | Sox Family | Tbx Family | |||||
| Human BMP Family | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||
NOTCH3
- Notch3 activation retains mammary luminal cell in a nonproliferative state [10]
NOTCH4
Notch Ligands
- JAG1
- JAG2
- DLL1
- DLL3
- DLL4
Functions
Developmental patterning signal.
Early Development
Blastocyst
| Mouse Blastocyst (32 cell stage) Fate | |
|---|---|
| Inner cells | Outer cells |
| Angiomotin (Amot) phosphorylation at adherens junctions | Amot sequestered by cell polarization from basolateral adherens junctions |
| Hippo active | Hippo inactive |
| Notch inactive | Notch active |
| Cdx2 not expressed | Cdx2 expressed |
| ICM - inner cell mass fate | TE - trophectoderm fate |
|
Hippo[11](TEAD4) and Notch[12](Cdx2) together appear regulate early blastocyst fate development. | |
Neural
Spinal Cord Development
Model of the embryonic rostro-caudal gradient of neurogenesis along the chicken spinal cord from the stem zone to the neurogenic neural tube summarising how DELTA-NOTCH signalling may be involved in these processes.[13]
|
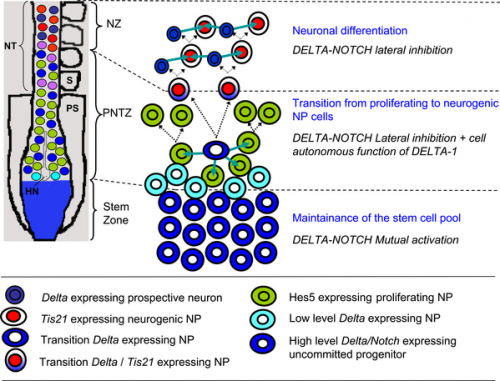
|
Hippocampus
Notch1 deficiency in postnatal neural progenitor cells in the dentate gyrus leads to emotional and cognitive impairment[14]
- "The latest evidence shows that Notch1 also plays a critical role in synaptic plasticity in mature hippocampal neurons. So far, deeper insights into the function of Notch1 signaling during the different steps of adult neurogenesis are still lacking, and the mechanisms by which Notch1 dysfunction is associated with brain disorders are also poorly understood. In the current study, we found that Notch1 was highly expressed in the adult-born immature neurons in the hippocampal dentate gyrus. Using a genetic approach to selectively ablate Notch1 signaling in late immature precursors in the postnatal hippocampus by cross-breeding doublecortin (DCX)+ neuron-specific proopiomelanocortin (POMC)-α Cre mice with floxed Notch1 mice, we demonstrated a previously unreported pivotal role of Notch1 signaling in survival and function of adult newborn neurons in the dentate gyrus. Moreover, behavioral and functional studies demonstrated that POMC-Notch1-/- mutant mice showed anxiety and depressive-like behavior with impaired synaptic transmission properties in the dentate gyrus. Finally, our mechanistic study showed significantly compromised phosphorylation of cAMP response element-binding protein (CREB) in Notch1 mutants, suggesting that the dysfunction of Notch1 mutants is associated with the disrupted pCREB signaling in postnatally generated immature neurons in the dentate gyrus."
- Links: Hippocampus Development
Endoderm Development
Endoderm differentiates to form the respiratory airway epithelium and glands. This epithelium is continuously replaced through life from a basal cell pool of undifferentiated airway progenitors. A recent study[5] has shown that the progenitor pool is regulated by the Notch3-Jagged signaling pathway. The mechanism appears dependent upon the availability of Jag1 and Jag2 (generating parabasal cells) that later activates Notch1 and Notch2 leading to a secretory-multiciliated cell fate.
- L;inks: Respiratory System Development
Mesoderm Development
Blood Vessel Development
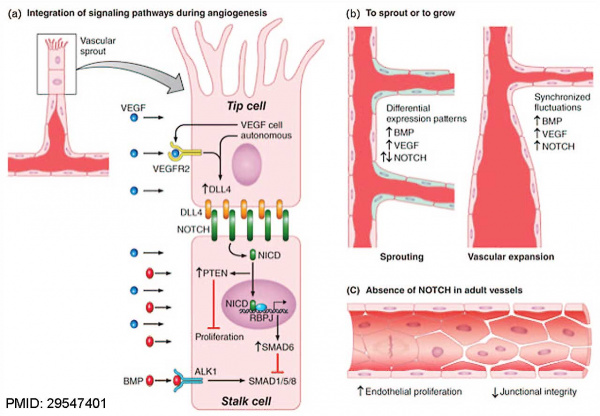
NOTCH Signaling in the EndotheliumPubmedParser error: Invalid PMID, please check. (PMID: 29547401PMID29547401)
(a) A vascular sprout is characterized by a leading ‘tip’ cell followed by ‘stalk’ cells.
(b) Regulation of new sprouting during vascular expansion depends on integration of BMP signaling, NOTCH signaling and VEGF signaling.
(c) In adult vessels, NOTCH is responsible for maintaining endothelial quiescence and junctional integrity.
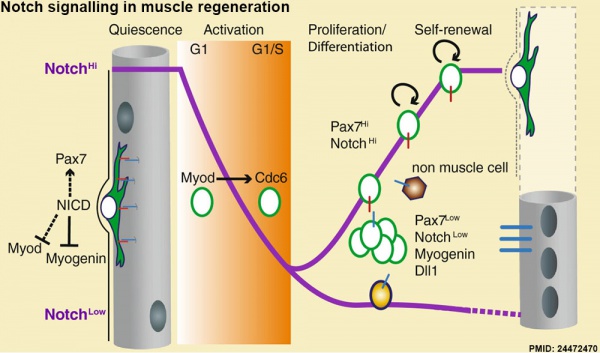
Muscle Regeneration
Notch signalling in muscle regeneration[15]
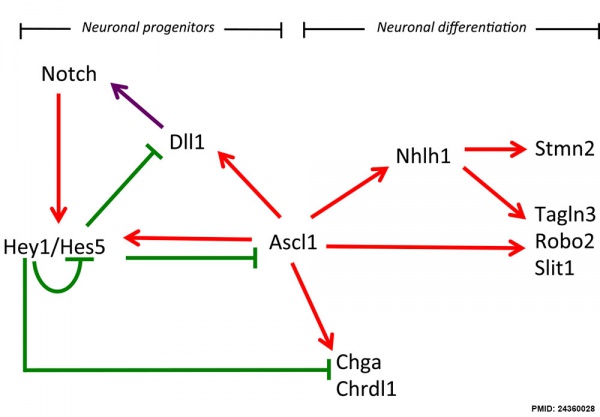
Hypothalamus Development
Hypothalamus Development Gene Interaction Model[16]
- Links: Hypothalamus Development
Abnormalities
Alagille Syndrome
Mutations in the human homolog of Jagged-1 (JAG1) located on chromosome 20p12 cause Alagille Syndrome. Abnormalities are seen in gastrointestinal (liver cholestasis), cardiac (heart), renal (kidney), skeletal, ocular, and facial systems.
- Links: Alagille Syndrome
References
- ↑ 1.0 1.1 Tien AC, Rajan A & Bellen HJ. (2009). A Notch updated. J. Cell Biol. , 184, 621-9. PMID: 19255248 DOI.
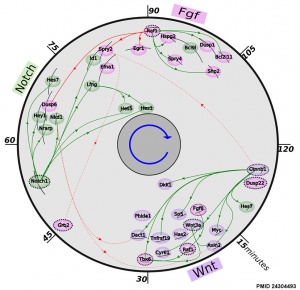
- ↑ 2.0 2.1 Fongang B & Kudlicki A. (2013). The precise timeline of transcriptional regulation reveals causation in mouse somitogenesis network. BMC Dev. Biol. , 13, 42. PMID: 24304493 DOI.
- ↑ Menchero S, Rollan I, Lopez-Izquierdo A, Andreu MJ, Sainz de Aja J, Kang M, Adan J, Benedito R, Rayon T, Hadjantonakis AK & Manzanares M. (2019). Transitions in cell potency during early mouse development are driven by Notch. Elife , 8, . PMID: 30958266 DOI.
- ↑ Hunter GL, Hadjivasiliou Z, Bonin H, He L, Perrimon N, Charras G & Baum B. (2016). Coordinated control of Notch/Delta signalling and cell cycle progression drives lateral inhibition-mediated tissue patterning. Development , 143, 2305-10. PMID: 27226324 DOI.
- ↑ 5.0 5.1 Mori M, Mahoney JE, Stupnikov MR, Paez-Cortez JR, Szymaniak AD, Varelas X, Herrick DB, Schwob J, Zhang H & Cardoso WV. (2015). Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development , 142, 258-67. PMID: 25564622 DOI.
- ↑ Mayeuf-Louchart A, Lagha M, Danckaert A, Rocancourt D, Relaix F, Vincent SD & Buckingham M. (2014). Notch regulation of myogenic versus endothelial fates of cells that migrate from the somite to the limb. Proc. Natl. Acad. Sci. U.S.A. , 111, 8844-9. PMID: 24927569 DOI.
- ↑ Schrauwen I & Van Camp G. (2010). The etiology of otosclerosis: a combination of genes and environment. Laryngoscope , 120, 1195-202. PMID: 20513039 DOI.
- ↑ 8.0 8.1 Mead TJ & Yutzey KE. (2009). Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc. Natl. Acad. Sci. U.S.A. , 106, 14420-5. PMID: 19590010 DOI.
- ↑ Schuster-Gossler K, Harris B, Johnson KR, Serth J & Gossler A. (2009). Notch signalling in the paraxial mesoderm is most sensitive to reduced Pofut1 levels during early mouse development. BMC Dev. Biol. , 9, 6. PMID: 19161597 DOI.
- ↑ Lafkas D, Rodilla V, Huyghe M, Mourao L, Kiaris H & Fre S. (2013). Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J. Cell Biol. , 203, 47-56. PMID: 24100291 DOI.
- ↑ Sasaki H. (2015). Position- and polarity-dependent Hippo signaling regulates cell fates in preimplantation mouse embryos. Semin. Cell Dev. Biol. , 47-48, 80-7. PMID: 25986053 DOI.
- ↑ Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Cañon S, Sasaki H, Hadjantonakis AK, de la Pompa JL, Rossant J & Manzanares M. (2014). Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev. Cell , 30, 410-22. PMID: 25127056 DOI.
- ↑ Hämmerle B & Tejedor FJ. (2007). A novel function of DELTA-NOTCH signalling mediates the transition from proliferation to neurogenesis in neural progenitor cells. PLoS ONE , 2, e1169. PMID: 18000541 DOI.
- ↑ Feng S, Shi T, Qiu J, Yang H, Wu Y, Zhou W, Wang W & Wu H. (2017). Notch1 deficiency in postnatal neural progenitor cells in the dentate gyrus leads to emotional and cognitive impairment. FASEB J. , 31, 4347-4358. PMID: 28611114 DOI.
- ↑ Mourikis P & Tajbakhsh S. (2014). Distinct contextual roles for Notch signalling in skeletal muscle stem cells. BMC Dev. Biol. , 14, 2. PMID: 24472470 DOI.
- ↑ Ratié L, Ware M, Barloy-Hubler F, Romé H, Gicquel I, Dubourg C, David V & Dupé V. (2013). Novel genes upregulated when NOTCH signalling is disrupted during hypothalamic development. Neural Dev , 8, 25. PMID: 24360028 DOI.
Reviews
Reichrath J & Reichrath S. (2020). A Snapshot of the Molecular Biology of Notch Signaling: Challenges and Promises. Adv. Exp. Med. Biol. , 1227, 1-7. PMID: 32072495 DOI.
Amelio I, Grespi F, Annicchiarico-Petruzzelli M & Melino G. (2012). p63 the guardian of human reproduction. Cell Cycle , 11, 4545-51. PMID: 23165243 DOI.
Roemer K. (2012). Notch and the p53 clan of transcription factors. Adv. Exp. Med. Biol. , 727, 223-40. PMID: 22399351 DOI.
Andersen P, Uosaki H, Shenje LT & Kwon C. (2012). Non-canonical Notch signaling: emerging role and mechanism. Trends Cell Biol. , 22, 257-65. PMID: 22397947 DOI.
Andersson ER, Sandberg R & Lendahl U. (2011). Notch signaling: simplicity in design, versatility in function. Development , 138, 3593-612. PMID: 21828089 DOI.
Articles
Search Pubmed
Search Bookshelf Notch
Search Pubmed Now: Notch Signaling
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- OMIM - NOTCH 1
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2026, February 28) Embryology Developmental Signals - Notch. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Developmental_Signals_-_Notch
- © Dr Mark Hill 2026, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G