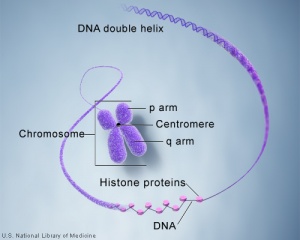
Molecular Development - Epigenetics
| Embryology - 28 Feb 2026 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
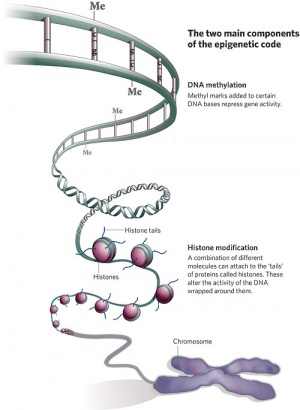
In terms of molecular mechanisms, the field of epigenetics has begun to florish with some recent important findings. Epigenetics as the name implies, is the inheritance mechanisms that lie outside the DNA sequence of our genes and include DNA methylation, histone modification, and those of the microRNA machinery.
One of the initial discoveries was the effects of DNA methylation upon gene expression and then modifications of nucleosomal histones. DNA methylation is usually associated with 5-methylcytosine (m5C) and leads to transcriptional silencing in vertebrates. Epigenetic modifications can be transmitted from one cell generation to the next (mitotic inheritance) and can also be transmitted down organismal generations (meiotic inheritance). Recently the term “methylome” has been coined to refer to the methylation profile of the whole genome.
Molecular mechanisms of development is an exciting research area and requires a variety of different skills. This page introduces only a few examples and should give you a feel for the topic. Note that each section of system notes has a page covering molecular development in that system.
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Development Epigenetics | Epigenetics | Oocyte Epigenetics | Spermatozoa Epigenetics |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
DNA Methylation
Enzymes that lead to DNA methylation are described as methyltransferases (DNMTs) and fall into two categories.
- DNMT1 - copies the pattern of DNA methylation during cell replication (methylation maintenance).
- DNMT3a and DNMT3b - are responsible for the de novo DNA methylation.
DNA Methylation Changes with Age
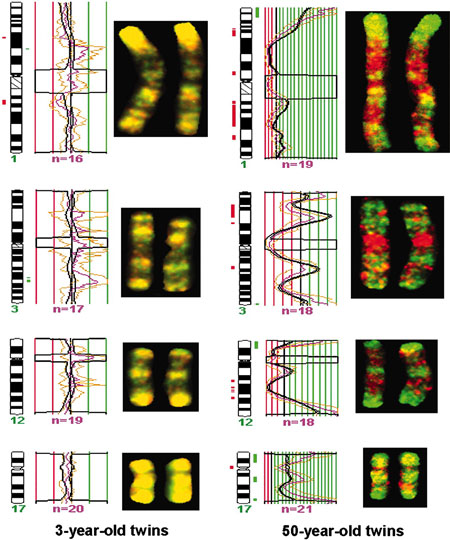
|
| DNA Methylation, young and old monozygous twins.[13] |
Developmental Methylation Changes
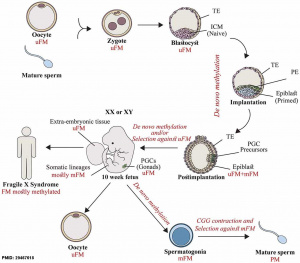
Within the embryonic genome DNA methylation occurs at regions of cytosine residues followed by guanines (CpG) and is a main epigenetic mechanism regulating early gene expression and later genomic imprinting, see review.[14] There are extensive changes in DNA methylation state that occur during development and relate to imprinted genes, the total number of genes imprinted in currently unknown, with 100 identified imprinted genes in the mouse and about 50 known in the human. This imprinting will also differ between the embryo and the placenta.[15]
Overall a developmental demethylation is followed by an eventual remethylation of about 70% of all CpGs, except for the primordial germ cell population. The primordial germ cells will form the germ line cell population in the embryo gonad. These cells appear to undergo epigenetic reprogramming through an independent genome-wide of erasure of imprints and epimutations through cytidine deaminases.[16]
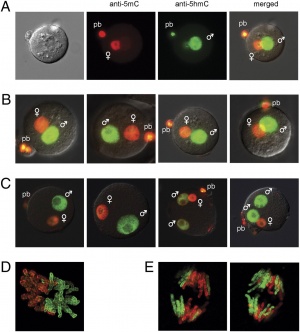
The zygote initially has a male and female pronucleus that will fuse to form the diploid nucleus. Each of the male and female pronuclei have different patterns of methylation and undergo different demethylation processes. The male pronucleus undergoes an extensive loss of DNA methylation, with imprinted genes resisting this process. The female pronucleus has less active demethylation requiring several mitotic rounds to complete this process.[14]
Primordial Germ Cells
|
Mouse primordial germ cell DNA methylation[17]
Demethylation
|
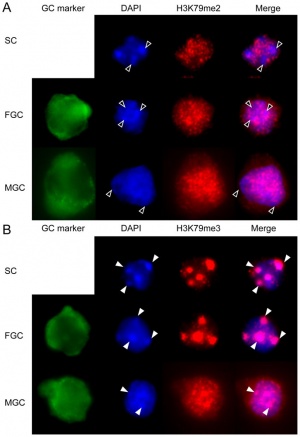
|
DNA Demethylation
Activation-Induced cytidine Deaminase
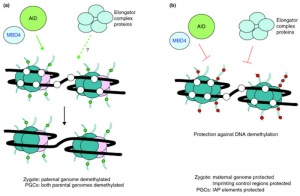
Activation-Induced cytidine Deaminase (AID) is an enzyme required for demethylation (removal of CpG methylation). Within the genome, DNA methylation is associated with epigenetic mechanisms and occurs at cytosine residues that are followed by guanines.[14] This enzyme is also found expressed in primordial germ cells.
Histone Modification
Histones are a family of proteins involved in the bundling of genomic DNA into chromatin. The unit size of chromatin DNA + associated histones is described as the nucleosome. A group of histone modifying enzymes can modify histone protein NH2-terminal tails by: acetylation, methylation, phosphorylation, sumoylation, or ubiquitination. These modifications of histone proteins can in turn determine the accessibility of the DNA to the transcription machinery.
Histone Acetylation
The lysine residues on histone tails can have acetyl groups either removed (by histone deacetyltransferases, HDACs) or added (by histone acetyl transferases, HATs).
- Normally the lysine residues on histone tails bear a positive charge that can bind negatively charged DNA to form a condensed structure with low transcriptional activity.
- Histone acetylation removes these positive charges allowing a less condensed structure with higher transcriptional activity.
Potential Imprinted Genes
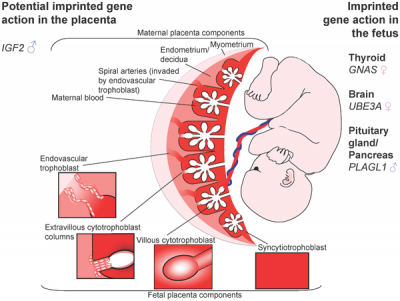
|
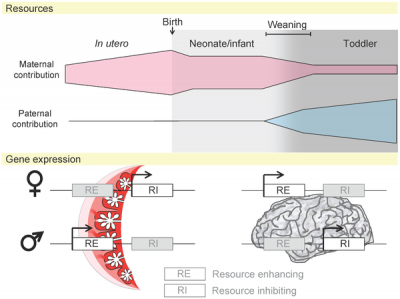
|
| Placenta potential imprinted genes[15] | Maternal and paternal resource allocation[15] |
X Inactivation
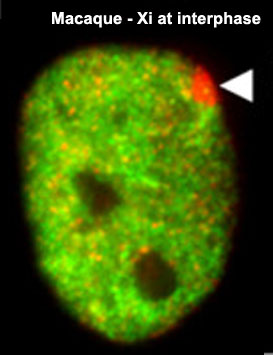
As X inactivation is not an encoded genetic function and occurs randomly throughout female tissues, this represents a form of epigenetics. The presence in females of two X chromosome raises the issue of gene dosage, in the case of mammals this is regulated by inactivating one of the X chromosomes. To balance expression with the autosomal chromosomes the dosage imbalance is then adjusted by doubling expression of X-linked genes in both sexes. The process of inactivation relies on the Xist RNA, a 17 kb non-coding RNA, which accumulates on the future inactive X (Xi) chromosome.
- Links: X Inactivation
References
- ↑ Qiu J. (2006). Epigenetics: unfinished symphony. Nature , 441, 143-5. PMID: 16688142 DOI.
- ↑ 2.0 2.1 Mor-Shaked H & Eiges R. (2018). Reevaluation ofFMR1Hypermethylation Timing in Fragile X Syndrome. Front Mol Neurosci , 11, 31. PMID: 29467618 DOI.
- ↑ Iqbal K, Jin SG, Pfeifer GP & Szabó PE. (2011). Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. , 108, 3642-7. PMID: 21321204 DOI.
- ↑ Ryan CP & Kuzawa CW. (2020). Germline epigenetic inheritance: Challenges and opportunities for linking human paternal experience with offspring biology and health. Evol. Anthropol. , , . PMID: 32196832 DOI.
- ↑ Arthur-Farraj P & Moyon S. (2020). DNA methylation in Schwann cells and in oligodendrocytes. Glia , , . PMID: 31958184 DOI.
- ↑ Sendžikaitė G & Kelsey G. (2019). The role and mechanisms of DNA methylation in the oocyte. Essays Biochem. , , . PMID: 31782490 DOI.
- ↑ Velker BAM, Denomme MM, Krafty RT & Mann MRW. (2017). Maintenance ofMestimprinted methylation in blastocyst-stage mouse embryos is less stable than other imprinted loci following superovulation or embryo culture. Environ Epigenet , 3, dvx015. PMID: 29492315 DOI.
- ↑ Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, Jin X, Shi X, Liu P, Wang X, Wang W, Wei Y, Li X, Guo F, Wu X, Fan X, Yong J, Wen L, Xie SX, Tang F & Qiao J. (2014). The DNA methylation landscape of human early embryos. Nature , 511, 606-10. PMID: 25079557 DOI.
- ↑ Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K & Meissner A. (2014). DNA methylation dynamics of the human preimplantation embryo. Nature , 511, 611-5. PMID: 25079558 DOI.
- ↑ Chowdhury S, Erickson SW, MacLeod SL, Cleves MA, Hu P, Karim MA & Hobbs CA. (2011). Maternal genome-wide DNA methylation patterns and congenital heart defects. PLoS ONE , 6, e16506. PMID: 21297937 DOI.
- ↑ PLoS Collection - Epigenetics 2010
- ↑ Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP & Daley GQ. (2010). Epigenetic memory in induced pluripotent stem cells. Nature , 467, 285-90. PMID: 20644535 DOI.
- ↑ Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C & Esteller M. (2005). Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. U.S.A. , 102, 10604-9. PMID: 16009939 DOI.
- ↑ 14.0 14.1 14.2 14.3 Sanz LA, Kota SK & Feil R. (2010). Genome-wide DNA demethylation in mammals. Genome Biol. , 11, 110. PMID: 20236475 DOI.
- ↑ 15.0 15.1 15.2 Frost JM & Moore GE. (2010). The importance of imprinting in the human placenta. PLoS Genet. , 6, e1001015. PMID: 20617174 DOI.
- ↑ Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE & Reik W. (2010). Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature , 463, 1101-5. PMID: 20098412 DOI.
- ↑ Abe M, Tsai SY, Jin SG, Pfeifer GP & Szabó PE. (2011). Sex-specific dynamics of global chromatin changes in fetal mouse germ cells. PLoS ONE , 6, e23848. PMID: 21886830 DOI.
- ↑ McLaughlin CR & Chadwick BP. (2011). Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biol. , 12, R37. PMID: 21489251 DOI.
Search Pubmed
June 2010 "epigenetics" - All (37097) Review (6256) Free Full Text (15778)
Search Pubmed Now: epigenetics
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2026, February 28) Embryology Molecular Development - Epigenetics. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Molecular_Development_-_Epigenetics
- © Dr Mark Hill 2026, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G