Developmental Signals - Hippo
| Embryology - 28 Feb 2026 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
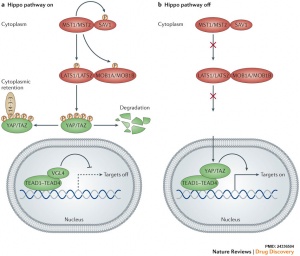
The Hippo (Hpo) pathway, first identified in Drosophila, controls organ size by regulating cell proliferation (inhibition) and apoptosis (induction). In contrast, the TOR signalling pathway regulates organ size by stimulating cell growth, thus increasing cell size.
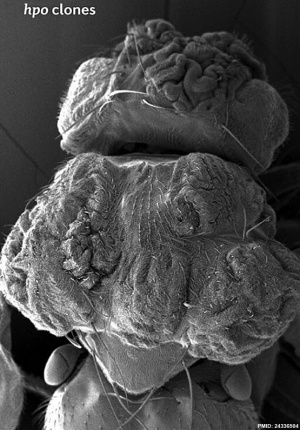
Hippo is a protein kinase cascade pathway, getting its name from the “hippopotamus”-like fly phenotype.
Current research suggests that this signaling pathways is involved with the first "patterning" decision cells make in the blastocyst, controlling trophectoderm vs inner cell mass decision making.
| Fly Phenotype (dorsal view head thorax SEM) | |
|---|---|
|

|
| Hippo-type (hpo) | Wild-type (WT) |
| Image source[1] |
| Factor Links: AMH | hCG | BMP | sonic hedgehog | bHLH | HOX | FGF | FOX | Hippo | LIM | Nanog | NGF | Nodal | Notch | PAX | retinoic acid | SIX | Slit2/Robo1 | SOX | TBX | TGF-beta | VEGF | WNT | Category:Molecular |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Embryo Hippo | Images | Embryo YAP | |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Early Development
Zygote
Maternally inherited Yes-associated protein (Yap), a co-activator of TEAD family transcription factors, plays a key role in activating embryonic transcription following fertilization in the mouse. Lysophosphatidic acid (LPA) in the mouse tubal fluid binds to its G-protein coupled receptor at the plasma membrane, and induces the activation of YAP by inhibiting LATS1/2. [5]
- Links: Zygote | Mouse Development
Blastocyst
Current research suggests that this signaling pathways is involved with the first "patterning" decision cells make in the blastocyst, controlling trophectoderm vs inner cell mass decision making.
| Mouse Blastocyst (32 cell stage) Fate | |
|---|---|
| Inner cells | Outer cells |
| Angiomotin (Amot) phosphorylation at adherens junctions | Amot sequestered by cell polarization from basolateral adherens junctions |
| Hippo active | Hippo inactive |
| Notch inactive | Notch active |
| Cdx2 not expressed | Cdx2 expressed |
| ICM - inner cell mass fate | TE - trophectoderm fate |
|
Hippo[6](TEAD4) and Notch[8](Cdx2) together appear regulate early blastocyst fate development. | |
- Links: blastocyst
Trophoblast
Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta PMAS "Defects in development and differentiation of the placenta are associated with various pregnancy disorders such as miscarriage, stillbirth, preeclampsia, and intrauterine growth restriction. However, our knowledge on critical regulators of human placentation is scarce. In the present study, we show that the Hippo signaling-dependent transcriptional coactivator YAP plays a pivotal role in the maintenance of proliferative trophoblasts, the epithelial cells of the placenta. By binding to the transcription factor TEAD4, YAP stimulates expression of genes promoting trophoblast stemness. Additionally, YAP–TEAD4 complexes actively repress genes associated with the differentiated syncytiotrophoblast, the hormone-secreting cell type of the human placenta. Hence, YAP orchestrates a complex developmental program ensuring growth and expansion of the human placenta. we herein show that YAP, the transcriptional coactivator of the Hippo signaling pathway, promotes maintenance of cytotrophoblast progenitors by different genomic mechanisms. Genetic or chemical manipulation of YAP in these cellular models revealed that it stimulates proliferation and expression of cell cycle regulators and stemness-associated genes, but inhibits cell fusion and production of syncytiotrophoblast (STB)-specific proteins, such as hCG and GDF15."
- Links: trophoblast
Bone
Hippo signaling pathway has recently been identified as a key regulator of osteoclast formation, see review.[9]
- Hippo signaling pathway regulatory molecules - RASSF2, NF2, MST1/2, SAV1, LATS1/2, MOB1, YAP, and TAZ.
- osteoclast differentiation - upon activation, MST and LAST, transcriptional co-activators YAP and TAZ bind to the members of the TEA domain (TEAD) family transcription factors
- regulate expression of downstream target genes connective tissue growth factor (CTGF/CCN2) and cysteine-rich protein 61 (CYR61/CCN1).
- RANKL-mediated signaling cascades including NF-κB, MAPKs, AP1, and NFATc1, Hippo-signaling molecules such as YAP/TAZ/TEAD complex, RASSF2, MST2, and Ajuba could also potentially modulate osteoclast differentiation and function.
- Links: bone
Neural
During early neural development radial glia guide the migration of differentiating neural cells to the developing cortical plate (cortex). A study has shown that elimination of CEP83, in radial glial progenitor cells, activates the mechanically sensitive yes-associated protein (YAP) and promotes the excessive proliferation of these progenitor cells.
- Links: radial glia | cortex
References
- ↑ 1.0 1.1 Johnson R & Halder G. (2014). The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov , 13, 63-79. PMID: 24336504 DOI.
- ↑ Piccolo FM, Kastan NR, Haremaki T, Tian Q, Laundos TL, De Santis R, Beaudoin AJ, Carroll TS, Luo JD, Gnedeva K, Etoc F, Hudspeth AJ & Brivanlou AH. (2022). Role of YAP in early ectodermal specification and a Huntington's Disease model of human neurulation. Elife , 11, . PMID: 35451959 DOI.
- ↑ Ishan M, Chen G, Yu W, Wang Z, Giovannini M, Cao X & Liu HX. (2021). Deletion of Nf2 in neural crest-derived tongue mesenchyme alters tongue shape and size, Hippo signalling and cell proliferation in a region- and stage-specific manner. Cell Prolif , 54, e13144. PMID: 34697858 DOI.
- ↑ Cho YS, Li S, Wang X, Zhu J, Zhuo S, Han Y, Yue T, Yang Y & Jiang J. (2020). CDK7 regulates organ size and tumor growth by safeguarding the Hippo pathway effector Yki/Yap/Taz in the nucleus. Genes Dev. , 34, 53-71. PMID: 31857346 DOI.
- ↑ 5.0 5.1 Yu C, Ji SY, Dang YJ, Sha QQ, Yuan YF, Zhou JJ, Yan LY, Qiao J, Tang F & Fan HY. (2016). Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res. , 26, 275-87. PMID: 26902285 DOI.
- ↑ 6.0 6.1 Sasaki H. (2015). Position- and polarity-dependent Hippo signaling regulates cell fates in preimplantation mouse embryos. Semin. Cell Dev. Biol. , 47-48, 80-7. PMID: 25986053 DOI.
- ↑ Porazinski S, Wang H, Asaoka Y, Behrndt M, Miyamoto T, Morita H, Hata S, Sasaki T, Krens SFG, Osada Y, Asaka S, Momoi A, Linton S, Miesfeld JB, Link BA, Senga T, Shimizu N, Nagase H, Matsuura S, Bagby S, Kondoh H, Nishina H, Heisenberg CP & Furutani-Seiki M. (2015). YAP is essential for tissue tension to ensure vertebrate 3D body shape. Nature , 521, 217-221. PMID: 25778702 DOI.
- ↑ Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Cañon S, Sasaki H, Hadjantonakis AK, de la Pompa JL, Rossant J & Manzanares M. (2014). Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev. Cell , 30, 410-22. PMID: 25127056 DOI.
- ↑ Yang W, Han W, Qin A, Wang Z, Xu J & Qian Y. (2018). The emerging role of Hippo signaling pathway in regulating osteoclast formation. J. Cell. Physiol. , 233, 4606-4617. PMID: 29219182 DOI.
Reviews
Irvine KD & Harvey KF. (2015). Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harb Perspect Biol , 7, . PMID: 26032720 DOI.
Tumaneng K, Russell RC & Guan KL. (2012). Organ size control by Hippo and TOR pathways. Curr. Biol. , 22, R368-79. PMID: 22575479 DOI.
Zhao B, Tumaneng K & Guan KL. (2011). The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. , 13, 877-83. PMID: 21808241 DOI.
Kango-Singh M & Singh A. (2009). Regulation of organ size: insights from the Drosophila Hippo signaling pathway. Dev. Dyn. , 238, 1627-37. PMID: 19517570 DOI.
Articles
Bessonnard S, Mesnard D & Constam DB. (2015). PC7 and the related proteases Furin and Pace4 regulate E-cadherin function during blastocyst formation. J. Cell Biol. , 210, 1185-97. PMID: 26416966 DOI.
Yuan H, Liu H, Liu Z, Zhu D, Amos CI, Fang S, Lee JE & Wei Q. (2015). Genetic variants in Hippo pathway genes YAP1, TEAD1 and TEAD4 are associated with melanoma-specific survival. Int. J. Cancer , 137, 638-45. PMID: 25628125 DOI.
Clattenburg L, Wigerius M, Qi J, Rainey JK, Rourke JL, Muruganandan S, Sinal CJ & Fawcett JP. (2015). NOS1AP Functionally Associates with YAP To Regulate Hippo Signaling. Mol. Cell. Biol. , 35, 2265-77. PMID: 25918243 DOI.
Search Pubmed
Search Bookshelf Hippo
Search Pubmed Now: Hippo
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2026, February 28) Embryology Developmental Signals - Hippo. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Developmental_Signals_-_Hippo
- © Dr Mark Hill 2026, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G
