| Educational Use Only - Embryology is an educational resource for learning concepts in embryological development, no clinical information is provided and content should not be used for any other purpose.
|
Introduction

Trisomy 21 (Down Syndrome) Male Karyotype
The table below shows the correlation of maternal age (mother's age) and the potential risk of human genetic abnormalities in children.
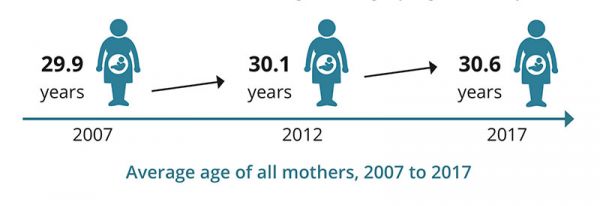
Australian Average Maternal Age Change
The first column shows maternal age, the second column shows the most common human chromosomal abnormality, trisomy 21 (Down syndrome), the third column shows all chromosomal abnormalities. The data below are from papers published in the 1980's.[1][2][3]
Interestingly, recent studies suggest that increasing paternal age (father's age) can also have affects on childhood mortality[4] and neurodevelopmental outcomes.[5]
The Oocyte Mosaicism Selection theory[6] suggests that "the incidence of trisomy 21 mosaicism in a cohort of normal fetal ovarian samples, indicating that an accumulation of trisomy 21 germ cells does indeed take place during fetal oogenesis, i.e., from the first to the second trimester of pregnancy. We presume that this accumulation of trisomy 21 (T21) cells is caused by their delay in maturation and lagging behind the normal cells. We further presume that this trend continues during the third trimester of pregnancy and postnatally, up until ovulation, thereby explaining the maternal age effect in Down syndrome." A similar selection model and ageing has been suggested for Trisomy 13.[7]
Age Table
Genetic Risk Maternal Age
| Age of Mother
|
Risk of Trisomy 21
|
Risk of Any Chromosomal Abnormality
|
| 20
|
1 in 1667
|
1 in 526
|
| 21
|
1 in 1667
|
1 in 526
|
| 22
|
1 in 1429
|
1 in 500
|
| 23
|
1 in 1429
|
1 in 500
|
| 24
|
1 in 1250
|
1 in 476
|
| 25
|
1 in 1250
|
1 in 476
|
| 26
|
1 in 1176
|
1 in 476
|
| 27
|
1 in 1111
|
1 in 455
|
| 28
|
1 in 1053
|
1 in 435
|
| 29
|
1 in 1000
|
1 in 417
|
| 30
|
1 in 952
|
1 in 384
|
| 31
|
1 in 909
|
1 in 384
|
| 32
|
1 in 769
|
1 in 323
|
| 33
|
1 in 625
|
1 in 286
|
| 34
|
1 in 500
|
1 in 238
|
| 35
|
1 in 385
|
1 in 192
|
| 36
|
1 in 294
|
1 in 156
|
| 37
|
1 in 227
|
1 in 127
|
| 38
|
1 in 175
|
1 in 102
|
| 39
|
1 in 137
|
1 in 83
|
| 40
|
1 in 106
|
1 in 66
|
| 41
|
1 in 82
|
1 in 53
|
| 42
|
1 in 64
|
1 in 42
|
| 43
|
1 in 50
|
1 in 33
|
| 44
|
1 in 38
|
1 in 26
|
| 45
|
1 in 30
|
1 in 21
|
| 46
|
1 in 23
|
1 in 16
|
| 47
|
1 in 18
|
1 in 13
|
| 48
|
1 in 14
|
1 in 10
|
| 49
|
1 in 11
|
1 in 8
|
| Table Data[1][2][3]
|
| Genetic risk maternal age
|
| Educational Use Only - Embryology is an educational resource for learning concepts in embryological development, no clinical information is provided and content should not be used for any other purpose.
|
|
Genetic Risk Maternal Age
| Age of Mother
|
Risk of Trisomy 21
|
Risk of Any Chromosomal Abnormality
|
| 20
|
1 in 1667
|
1 in 526
|
| 21
|
1 in 1667
|
1 in 526
|
| 22
|
1 in 1429
|
1 in 500
|
| 23
|
1 in 1429
|
1 in 500
|
| 24
|
1 in 1250
|
1 in 476
|
| 25
|
1 in 1250
|
1 in 476
|
| 26
|
1 in 1176
|
1 in 476
|
| 27
|
1 in 1111
|
1 in 455
|
| 28
|
1 in 1053
|
1 in 435
|
| 29
|
1 in 1000
|
1 in 417
|
| 30
|
1 in 952
|
1 in 384
|
| 31
|
1 in 909
|
1 in 384
|
| 32
|
1 in 769
|
1 in 323
|
| 33
|
1 in 625
|
1 in 286
|
| 34
|
1 in 500
|
1 in 238
|
| 35
|
1 in 385
|
1 in 192
|
| 36
|
1 in 294
|
1 in 156
|
| 37
|
1 in 227
|
1 in 127
|
| 38
|
1 in 175
|
1 in 102
|
| 39
|
1 in 137
|
1 in 83
|
| 40
|
1 in 106
|
1 in 66
|
| 41
|
1 in 82
|
1 in 53
|
| 42
|
1 in 64
|
1 in 42
|
| 43
|
1 in 50
|
1 in 33
|
| 44
|
1 in 38
|
1 in 26
|
| 45
|
1 in 30
|
1 in 21
|
| 46
|
1 in 23
|
1 in 16
|
| 47
|
1 in 18
|
1 in 13
|
| 48
|
1 in 14
|
1 in 10
|
| 49
|
1 in 11
|
1 in 8
|
| Table Data[1][2][3]
|
|
Some Recent Findings
- Preimplantation genetic testing for aneuploidy: a comparison of live birth rates in patients with recurrent pregnancy loss due to embryonic aneuploidy or recurrent implantation failure Sato T, Sugiura-Ogasawara M, Ozawa F, Yamamoto T, Kato T, Kurahashi H, Kuroda T, Aoyama N, Kato K, Kobayashi R, Fukuda A, Utsunomiya T, Kuwahara A, Saito H, Takeshita T & Irahara M. (2019). Preimplantation genetic testing for aneuploidy: a comparison of live birth rates in patients with recurrent pregnancy loss due to embryonic aneuploidy or recurrent implantation failure. Hum. Reprod. , , . PMID: 31811307 DOI. "Can preimplantation genetic testing for aneuploidy (PGT-A) improve the live birth rate and reduce the miscarriage rate in patients with recurrent pregnancy loss (RPL) caused by an abnormal embryonic karyotype and recurrent implantation failure (RIF)? ...A large portion of pregnancy losses in the RPL group might be due to aneuploidy, since PGT-A reduced the overall incidence of pregnancy loss in these patients. Although PGT-A did not improve the live birth rate per patient, it did have the advantage of reducing the number of embryo transfers required to achieve a similar number live births compared with those not undergoing PGT-A."
- Advanced maternal age and risk of non-chromosomal anomalies: data from a tertiary referral hospital in Turkey[8] "The purpose of this study is to determine if there is a relationship between non-chromosomal fetal anomalies of various organ systems and advanced maternal age. MATERIALS AND METHOD: This study was conducted in 387 women aged 20-53 years who underwent fetal karyotype testing due to positive prenatal test results or advanced maternal age at the Kanuni Sultan Süleyman Training and Research Hospital between September 2011 and March 2015. Fetuses with chromosomal anomalies were excluded from the study. The relationship between non-chromosomal anomalies and maternal age of women aged <35 or ≥35 years was studied. RESULTS: More than 80% (81.7%) of non-chromosomal anomalies were detected in patients aged <35 years, and 18.3% were found in those ≥35 years. There were no statistically significant differences found between the incidence of non-chromosomal anomalies in women aged over 35 years and those under 35 years. When congenital major anomalies were evaluated with respect to various organ systems, the risk of musculo-skeletal system anomalies decreased with advancing maternal age. However, there was no statistically significant difference between the <35 and ≥35-year age groups in the incidence of central nervous system, craniofacial, cardiac, gastrointestinal system, urogenital, respiratory, and limb anomalies. CONCLUSION: The incidence of non-chromosomal anomalies does not increase in fetuses of pregnant women aged over 35 years, in contrast to chromosomal anomalies."
- Outcomes of Women Delivering at Very Advanced Maternal Age[9] "Retrospective cohort study using the Texas Public Use Data File, years 2013-2014 (96,879 deliveries). Maternal age was a three-level variable: 35-39 (referent), 40-44, and 45-59 years (VAMA). Adjusted risk ratios (aRRs) for the two older age groups for various obstetrical and nonobstetrical complications were calculated from log-binomial regression models. RESULTS: The sample consisted of 96,879 deliveries. In univariate analyses, a higher frequency (p < 0.05) of gestational diabetes, pregestational diabetes, chronic hypertension, pregnancy related hypertensive disorders, multiple gestation, oligohydramnios, polyhydramnios, placenta previa, postpartum hemorrhage, small for gestational age, intrauterine fetal death, and length of stay were noted in the two older maternal age groups compared to the youngest maternal age group. Multiple gestations were noted to be more frequent in the two older groups. The risk of the following outcomes was approximately doubled in VAMA women compared to the referent (all statistically significant): small for gestational age (aRR = 1.92), stillbirth (aRR = 2.12), and intrauterine fetal death (aRR = 1.96). CONCLUSIONS: This population-based study detected a dose-response association between maternal age and the risk of multiple maternal and fetal complications."
- Epidemiology of chromosomal trisomies in the East of Ireland[10] "Chromosomal trisomies are associated with advancing maternal age. In Ireland, information on the total prevalence and outcome of trisomy affected pregnancies is unavailable. This study aimed to ascertain more precise data on Trisomies 21, 18 and 13 in a large Irish region during the period 2011-2013. ...Over 90% of Trisomies 18/13 and 47% of Trisomy 21 were diagnosed prenatally; 61% of Trisomy 21 cases and nearly 30% of Trisomies 18/13 were live births; 38% all trisomy affected pregnancies ended in a termination. CONCLUSIONS: This study provides precise data on the total prevalence and outcome of trisomy affected pregnancies in the East of Ireland. Total prevalence rates were higher than previously reported. Prenatal diagnosis had a significant impact on outcome. These data provide a better basis for planning of services for live-born children affected by trisomy."
|
| More recent papers
|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
- This search now requires a manual link as the original PubMed extension has been disabled.
- The displayed list of references do not reflect any editorial selection of material based on content or relevance.
- References also appear on this list based upon the date of the actual page viewing.
References listed on the rest of the content page and the associated discussion page (listed under the publication year sub-headings) do include some editorial selection based upon both relevance and availability.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References
Search term: Maternal Age Abnormal Development | Maternal Age Genetic Abnormalities
|
| Older papers
|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
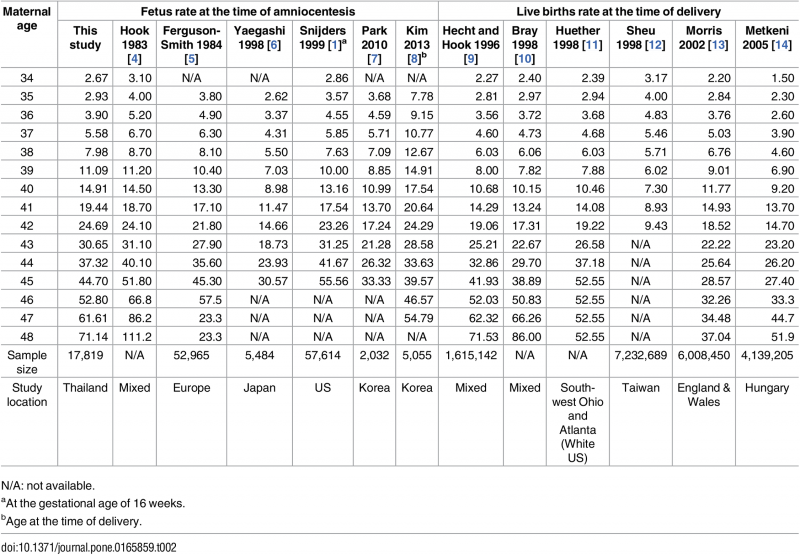
- Maternal Age-Specific Rates for Trisomy 21 and Common Autosomal Trisomies in Fetuses from a Single Diagnostic Center in Thailand[11] "To provide maternal age-specific rates for trisomy 21 (T21) and common autosomal trisomies (including trisomies 21, 18 and 13) in fetuses. We retrospectively reviewed prenatal cytogenetic results obtained between 1990 and 2009 in Songklanagarind Hospital, a university teaching hospital, in southern Thailand. Maternal age-specific rates of T21 and common autosomal trisomies were established using different regression models, from which only the fittest models were used for the study. A total of 17,819 records were included in the statistical analysis. The fittest models for predicting rates of T21 and common autosomal trisomies were regression models with 2 parameters (Age and Age2). The rate of T21 ranged between 2.67 per 1,000 fetuses at the age of 34 and 71.06 per 1,000 at the age of 48. The rate of common autosomal trisomies ranged between 4.54 per 1,000 and 99.65 per 1,000 at the same ages. This report provides the first maternal age-specific rates for T21 and common autosomal trisomies fetuses in a Southeast Asian population and the largest case number of fetuses have ever been reported in Asians."
- Sex ratios among infants with birth defects, National Birth Defects Prevention Study, 1997-2009[12] "A small number of population-based studies have examined sex differences among infants with birth defects. The highest elevations in sex ratios (i.e., male preponderance) among isolated non-cardiac defects were for craniosynostosis (2.12), cleft lip with cleft palate (2.01), and cleft lip without cleft palate (1.78); the lowest sex ratios (female preponderance) were for choanal atresia (0.45), cloacal exstrophy (0.46), and holoprosencephaly (0.64). Among isolated cardiac defects, the highest sex ratios were for aortic stenosis (2.88), coarctation of the aorta (2.51), and d-transposition of the great arteries (2.34); the lowest were multiple ventricular septal defects (0.52), truncus arteriosus (0.63), and heterotaxia with congenital heart defect (0.64)."
|
Maternal Age Trisomy 21 Studies
Comparative data compiled by this study.[11]
Ireland
Data from a clinical data study of chromosomal trisomies in the East of Ireland (2011-2013).[10]
Total births 80,894 - 394 trisomy cases (prevalence rate 48.9/10,000)
Diagnosed prenatally
- 90+% trisomies 18/13
- 47% of Trisomy 21
Thailand
The following data is from a recent Thai study of maternal age and trisomies.[11]
Genetic Risk Maternal Age
| Age of Mother
|
Risk of Trisomy 21
|
Risk of Any Autosomal Trisomies
|
| Genetic Risk Maternal Age
|
| Age of Mother
|
Risk of Trisomy 21
|
Risk of Any Autosomal Trisomies
|
| 34
|
2.67 in 1,000
|
4.54 in 1,000
|
| 48
|
71.06 in 1,000
|
99.65 in 1,000
|
| Thai study of maternal age and trisomies.[11]
|
References
- ↑ 1.0 1.1 1.2 Hook EB. (1981). Rates of chromosome abnormalities at different maternal ages. Obstet Gynecol , 58, 282-5. PMID: 6455611
- ↑ 2.0 2.1 2.2 Hook EB, Cross PK & Schreinemachers DM. (1983). Chromosomal abnormality rates at amniocentesis and in live-born infants. JAMA , 249, 2034-8. PMID: 6220164
- ↑ 3.0 3.1 3.2 Schreinemachers DM, Cross PK & Hook EB. (1982). Rates of trisomies 21, 18, 13 and other chromosome abnormalities in about 20 000 prenatal studies compared with estimated rates in live births. Hum. Genet. , 61, 318-24. PMID: 6891368
- ↑ Zhu JL, Vestergaard M, Madsen KM & Olsen J. (2008). Paternal age and mortality in children. Eur. J. Epidemiol. , 23, 443-7. PMID: 18437509 DOI.
- ↑ Saha S, Barnett AG, Foldi C, Burne TH, Eyles DW, Buka SL & McGrath JJ. (2009). Advanced paternal age is associated with impaired neurocognitive outcomes during infancy and childhood. PLoS Med. , 6, e40. PMID: 19278291 DOI.
- ↑ Hultén MA, Öijerstedt L, Iwarsson E & Jonasson J. (2014). Maternal Germinal Trisomy 21 in Down Syndrome. J Clin Med , 3, 167-75. PMID: 26237255 DOI.
- ↑ Babay LÉ, Horányi D, Győrffy B & Nagy GR. (2019). Evidence for the Oocyte Mosaicism Selection model on the origin of Patau syndrome (trisomy 13). Acta Obstet Gynecol Scand , 98, 1558-1564. PMID: 31464342 DOI.
- ↑ Okmen Ozkan B, Köroğlu N, Turkgeldi LS, Cetin BA & Aslan H. (2019). Advanced maternal age and risk of non-chromosomal anomalies: data from a tertiary referral hospital in Turkey. J. Matern. Fetal. Neonatal. Med. , 32, 749-752. PMID: 28992718 DOI.
- ↑ Arya S, Mulla ZD & Plavsic SK. (2018). Outcomes of Women Delivering at Very Advanced Maternal Age. J Womens Health (Larchmt) , , . PMID: 30016194 DOI.
- ↑ 10.0 10.1 McDonnell R, Monteith C, Kennelly M, Martin A, Betts D, Delany V, Lynch SA, Coulter-Smith S, Sheehan S & Mahony R. (2017). Epidemiology of chromosomal trisomies in the East of Ireland. J Public Health (Oxf) , 39, e145-e151. PMID: 27591300 DOI.
- ↑ 11.0 11.1 11.2 11.3 Jaruthamsophon K, Sriplung H, Charalsawadi C & Limprasert P. (2016). Maternal Age-Specific Rates for Trisomy 21 and Common Autosomal Trisomies in Fetuses from a Single Diagnostic Center in Thailand. PLoS ONE , 11, e0165859. PMID: 27812158 DOI.
- ↑ Michalski AM, Richardson SD, Browne ML, Carmichael SL, Canfield MA, VanZutphen AR, Anderka MT, Marshall EG & Druschel CM. (2015). Sex ratios among infants with birth defects, National Birth Defects Prevention Study, 1997-2009. Am. J. Med. Genet. A , 167A, 1071-81. PMID: 25711982 DOI.
Articles
Goetzinger KR, Shanks AL, Odibo AO, Macones GA & Cahill AG. (2017). Advanced Maternal Age and the Risk of Major Congenital Anomalies. Am J Perinatol , 34, 217-222. PMID: 27398707 DOI.
This study aims to determine if advanced maternal age (AMA) is a risk factor for major congenital anomalies, in the absence of aneuploidy. ...Of 76,156 euploid fetuses, 2.4% (n = 1,804) were diagnosed with a major anomaly. There was a significant decrease in the incidence of major fetal anomalies with increasing maternal age until the threshold of age 35 (p < 0.001). Being AMA was significantly associated with an overall decreased risk for major fetal anomalies (adjusted odds ratio: 0.59, 95% confidence interval: 0.52-0.66). The subgroup analysis demonstrated similar results for women ≥ 40 years of age. Conclusion AMA is associated with an overall decreased risk for major anomalies. These findings may suggest that the "all or nothing" phenomenon plays a more robust role in embryonic development with advancing oocyte age, with anatomically normal fetuses being more likely to survive."
Hultén MA, Öijerstedt L, Iwarsson E & Jonasson J. (2014). Maternal Germinal Trisomy 21 in Down Syndrome. J Clin Med , 3, 167-75. PMID: 26237255 DOI.
"It has now been over 50 years since it was discovered that Down syndrome is caused by an extra chromosome 21, i.e., trisomy 21. In the interim, it has become clear that in the majority of cases, the extra chromosome is inherited from the mother, and there is, in this respect, a strong maternal age effect. Numerous investigations have been devoted to clarifying the underlying mechanism, most recently suggesting that this situation is exceedingly complex, involving both biological and environmental factors. On the other hand, it has also been proposed that germinal trisomy 21 mosaicism, arising during the very early stages of maternal oogenesis with accumulation of trisomy 21 germ cells during subsequent development, may be the main predisposing factor. We present data here on the incidence of trisomy 21 mosaicism in a cohort of normal fetal ovarian samples, indicating that an accumulation of trisomy 21 germ cells does indeed take place during fetal oogenesis, i.e., from the first to the second trimester of pregnancy. We presume that this accumulation of trisomy 21 (T21) cells is caused by their delay in maturation and lagging behind the normal cells. We further presume that this trend continues during the third trimester of pregnancy and postnatally, up until ovulation, thereby explaining the maternal age effect in Down syndrome."
Csermely G, Czeizel AE & Veszprémi B. (2015). Distribution of maternal age and birth order groups in cases with unclassified multiple congenital abnormalities according to the number of component abnormalities: a national population-based case-control study. Birth Defects Res. Part A Clin. Mol. Teratol. , 103, 67-75. PMID: 25224265 DOI.
- "The Hungarian Case-Control Surveillance of Congenital Abnormalities, 1980 to 1996, yielded a large population-based national data set with 22,843 malformed newborns or fetuses ("informative cases") included 1349 UMCA cases with their 2407 matched controls. Case-control comparison of maternal age and birth order was made for cases with UMCA, stratified by component numbers and their controls. In addition, 834 cases with Down syndrome were compared to 1432 matched controls. ...The well-known advanced maternal age with the higher risk for Down syndrome was confirmed. The findings of the study suggest that the young age of mothers associates with the higher risk of UMCA, in addition birth order 4 or more associates with the higher risk for UMCA with 2 and 3 component CAs."
Allen EG, Freeman SB, Druschel C, Hobbs CA, O'Leary LA, Romitti PA, Royle MH, Torfs CP & Sherman SL. (2009). Maternal age and risk for trisomy 21 assessed by the origin of chromosome nondisjunction: a report from the Atlanta and National Down Syndrome Projects. Hum. Genet. , 125, 41-52. PMID: 19050929 DOI.
- "We examined the association between maternal age and chromosome 21 nondisjunction by origin of the meiotic error. We analyzed data from two population-based, case-control studies: Atlanta Down Syndrome Project (1989-1999) and National Down Syndrome Project (2001-2004). Cases were live born infants with trisomy 21 and controls were infants without trisomy 21 delivered in the same geographical regions. We enrolled 1,215 of 1,881 eligible case families and 1,375 of 2,293 controls. We report four primary findings. First, the significant association between advanced maternal age and chromosome 21 nondisjunction was restricted to meiotic errors in the egg; the association was not observed in sperm or in post-zygotic mitotic errors. Second, advanced maternal age was significantly associated with both meiosis I (MI) and meiosis II (MII). For example, compared to mothers of controls, mothers of infants with trisomy 21 due to MI nondisjunction were 8.5 times more likely to be >or=40 years old than 20-24 years old at the birth of the index case (95% CI=5.6-12.9). Where nondisjunction occurred in MII, mothers were 15.1 times more likely to be >or=40 years (95% CI = 8.4-27.3). Third, the ratio of MI to MII errors differed by maternal age. The ratio was lower among women <19 years of age and those >or=40 years (2.1, 2.3, respectively) and higher in the middle age group (3.6). Lastly, we found no effect of grand-maternal age on the risk for maternal nondisjunction. This study emphasizes the complex association between advanced maternal age and nondisjunction of chromosome 21 during oogenesis."
Munné S, Chen S, Colls P, Garrisi J, Zheng X, Cekleniak N, Lenzi M, Hughes P, Fischer J, Garrisi M, Tomkin G & Cohen J. (2007). Maternal age, morphology, development and chromosome abnormalities in over 6000 cleavage-stage embryos. Reprod. Biomed. Online , 14, 628-34. PMID: 17509208 DOI.
- "Previous studies assessing the relationship between embryo development, maternal age and chromosome abnormalities were either small or analysed mostly embryos not suitable for replacement. The present study includes >6000 embryos, including many suitable for replacement. Embryos with the best morphology and development were 44% euploid in patients younger than 35, decreasing to 21% in patients 41 and older. The worst morphology group had only 30% normal embryos from patients younger than 35, and 12% in embryos from patients 41 and older. Thus morphological analysis was able to improve the population of normal embryos only from 30 to 44% in the best of cases. Regarding specific abnormalities, 20% of embryos were aneuploid, 32% aneuploid plus other abnormalities, and the rest had post-meiotic abnormalities. Of those, only aneuploidy increased with maternal age. There were no big differences in the frequency of chromosome abnormalities depending on patient indication, within a similar age group. In summary, previous trends detected in suboptimal embryos were also confirmed in the best embryos for replacement. Although dysmorphism and advanced maternal age are both related to chromosome abnormalities, these parameters can yield at most <50% euploid embryos, and other techniques such as preimplantation diagnosis are required to ensure that only euploid embryos are replaced."
Kuliev A, Cieslak J & Verlinsky Y. (2005). Frequency and distribution of chromosome abnormalities in human oocytes. Cytogenet. Genome Res. , 111, 193-8. PMID: 16192694 DOI.
- "It was previously shown that more than half of the human oocytes obtained from IVF patients of advanced reproductive age are aneuploid, due to meiosis I and meiosis II errors. The present paper further confirms that 61.8% of the oocytes tested by fluorescent probes specific for chromosomes 13, 16, 18, 21 and 22 are abnormal, representing predominantly chromatid errors, which are the major source of aneuploidy in the resulting embryos. Almost half of the oocytes with meiosis I errors (49.3%) are prone to sequential meiosis II errors, which may lead to aneuploidy rescue in 30.8% of the cases. Half of the detected aneuploidies (49.8%) are of complex nature with involvement of two or more chromosomes, or the same chromosome in both meiotic divisions. The aneuploidy rates for individual chromosomes are different, with a higher prevalence of chromosome 21 and 22 errors. The origin of aneuploidy for the individual chromosomes is also not random, with chromosome 16 and 22 errors originating more frequently in meiosis II, and chromosome 18, 13 and 21 errors in meiosis I. There is an age dependence not only for the overall frequency of aneuploidies, but also for each chromosome error, aneuploidies originating from meiosis I, meiosis II, and both meiosis I and meiosis II errors, as well as for different types of aneuploidies. The data further suggest the practical relevance of oocyte aneuploidy testing for detection and avoidance from transfer of the embryos deriving from aneuploid oocytes, which should contribute significantly to the pregnancy outcomes of IVF patients of advanced reproduction age."
Search PubMed
Search PubMed Now: Genetic risk maternal age
Terms
| Genetic Terms (expand to view)
|
genetic abnormalities | Molecular Development | meiosis | mitosis
- Alpha-Fetoprotein test (APF test) A prenatal test to measure the amount of a fetal protein in the mother's blood (or amniotic fluid). Abnormal amounts of the protein may indicate genetic or developmental problems in the fetus. Serum alpha-fetoprotein (AFP) is a fetal glycoprotein produced by the yolk sac and fetal liver. Low levels of AFP normally occur in the blood of a pregnant woman, high levels may indicate neural tube defects (spina bifida, anencephaly). (More? Alpha-Fetoprotein)
- anaphase B - Cell division term referring to the part of anaphase during which the poles of the mitotic spindle move apart. (More? mitosis)
- antisense - a sequence of DNA that is complementary usually to coding sequence of DNA or mRNA. Has been used experimentally to perturb or block gene expression. Also a mechanism that has been found to occur naturally as a regulatory mechanism.
- aneuploidy - Genetic term used to describe an abnormal number of chromosomes mainly (90%) due to chromosome malsegregation mechanisms in maternal meiosis I.
- autosomal inheritance - some hereditary diseases are described as autosomal which means that the disease is due to a DNA error in one of the 22 pairs that are not sex chromosomes. Both boys and girls can then inherit this error. If the error is in a sex chromosome, the inheritance is said to be sex-linked.
- base - another term for nucleotide (usually a t c g).
- base pair - Double stranded DNA has nucleotides A-T, C-G, paired by hydrogen bonds (2 for AT, 3 for GC). Note this means that GC is harder to separate that AT.
- cis-acting elements - DNA sequences that through transcription factors or other trans-acting elements or factors, regulate the expression of genes on the same chromosome.
- cohesin - a multi-protein subunit complex required to keep the sister chromatids together until their separation at anaphase (both in mitosis and meiosis), can also form rings that connect two DNA segments.
- copy number variation - (copy number variants,CNVs) a DNA segment of one kilobase (kb) or larger that is present at a variable copy number in comparison with a reference genome. (More? Nature)
- disomy - Genetic term referring to the presence of two chromosomes of a homologous pair in a cell, as in diploid. See chromosomal number genetic disorders uniparental disomy and aneuploidy. Humans have pairs usually formed by one chromosome from each parent.
- DNMT - DNA methyltransferase.
- DNA - DeoxyriboNucleic Acid. The genetic material found in mammalian chromosomes and mitochondria. Consisting of 4 nucleic acids (ATCG) that combine in a triptych (3 nucleotide codon) code for protein amino acids (3nt = 1aa).
- DNA duplex - double stranded base-paired DNA forming a helix.
- dominant inheritance - With autosomal dominant inheritance, there is an error in one of the 22 chromosome pairs. But the damaged gene dominates over the normal gene received from the other parent. If one of the parents has a disease caused by an autosomal dominant gene, all the children will have a 50 per cent risk of inheriting the dominant gene and a 50 per cent chance of not inheriting it. The children who do not inherit the damaged dominant gene will not themselves suffer from the disease, nor will they be able to pass the gene on to future children. This type of inheritance is present for example in Huntington's disease.
- Down Syndrome - The historic name used for Trisomy 21, named after the original identifier Down, J.L.H. in a 1866 paper (cited above).
- enhancer - A cis-regulatory sequence that can regulate levels of transcription from an adjacent promoter. Many tissue-specific enhancers can determine spatial patterns of gene expression in higher eukaryotes. Enhancers can act on promoters over many tens of kilobases of DNA and can be 5' or 3' to the promoter they regulate.
- exon - a block of protein encoding sequence of DNA in a gene. Many proteins are made of several exons "stitched" or spliced together by editing out non-coding (intron) sequences.
- fasta - a format for listing DNA sequence, where the first line has descritive information followed on the next line by the sequence without numbering.
- GC repeat - a string of GC sequence repeated several times. Also associated with GC expansion, a mutational process that may lead eventually to serious gene expression effects.
- gene - a sequence of DNA that encodes an individual protein.
- genetic code - the 3 nucleotide sequence that forms a codon for a single amino acid or stop. See the gene code.
- genome - the complete genetic information in the form of DNA available to a specific species.
- hairpin loop - a folding of RNA generated by base pairing making a "===()" structure, the end loop and or stem of this structure can then interact with proteins or other RNA.
- heteroplasmy - within a cell when more than one type of mitochondrial DNA (mtDNA) genome exists within the mitochondrion, or between mitochondria. Heteroplasmy can generate a pathogenic mutation, for example transfer RNA leucine (tRNALeu(UUR)) 3243A > G mutant can result in diabetes.
- HyperD - hypermethylated domain has a role in regulating gene expression and epigenomics. The early embryo has large alterations in methylation patterns and DNA modification. (More? PMID 1943996)
- HypoD - hypomethylated domain has a role in regulating gene expression and epigenomics. The early embryo has large alterations in methylation patterns and DNA modification. (More? PMID 1943996)
- igDMR - imprinted germline differentially methylated regions
- intron - a block of DNA within a gene not encoding a protein. Edited, spliced, out during transcription into mRNA. Originally thought not to contain any information, but more and more this appears not to be the case. Some intron sequences have been shown to regulate gene expression during development (eg c elegans, Lin 14)
- karyotype - [Greek, karyon = kernel or nucleus + typos= stamp] The chromosomal makeup of a cell. Karyotyping describes the clinical genetic test.
- meiosis I (MI) The first part of meiosis resulting in separation of homologous chromosomes, in humans producing two haploid cells (N chromosomes, 23), a reductional division.
- Meiosis I: Prophase I - Metaphase I - Anaphase I - Telophase I
- meiosis II - (MII) The second part of meiosis. In male human spermatogenesis, producing of four haploid cells (23 chromosomes, 1N) from the two haploid cells (23 chromosomes, 1N), each of the chromosomes consisting of two sister chromatids produced in meiosis I. In female human oogenesis, only a single haploid cell (23 chromosomes, 1N) is produced. Meiosis II: Prophase II - Metaphase II - Anaphase II - Telophase II
- mosaic autosome monosomy - genetic abnormality caused by embryonic fusion or loss of an autosome early in embryonic development, resulting in a subset of cells in the body having only one of a pair of autosomes. ICD-11 LD43.1 Mosaic monosomy of autosome
- mRNA - messenger, transcribed from DNA in the nucleus and in mitochondria. Is translated by the ribosome in the cytoplasm (or mitochondrial matrix). Intermediate step in gene expression. (DNA-> mRNA-> protein).
- mutation - any process which results in the alteration of the DNA sequence. Some conservative mutations may have no effect on the final amino acid encoded.
- nuchal translucency - (fetal nuchal-translucency thickness) An initial Trisomy 21 diagnostic ultrasound measurement in the fetal neck region carried out by trans-abdominal ultrasound at gestational age GA 10–14 weeks. Fetal sagittal section scan at a magnification that the fetus occupied at least 75% of the image. Measured is the maximum thickness of the subcutaneous translucency between the skin and the soft tissue overlying the cervical spine.
- Philadelphia chromosome - (Philadelphia translocation) Genetic term referring to a chromosomal abnormality resulting from a reciprocal translocation between chromosome 9 and 22 (t(9;22)(q34;q11)). This is associated with the disease chronic myelogenous leukemia (CML).
- ploidy - refers to the chromosomal genetic content of cells.
- promoter - A regulatory region a short distance upstream from the 5' end of a transcription start site that acts as the binding site for RNA polymerase II. A region of DNA to which RNA polymerase IIbinds in order to initiate transcription.
- point mutation - a change in a single nucleotide.
- recessive inheritance - With autosomal recessive inheritance, the diseased individual has inherited the same gene damage from both father and mother. The damage is found on both chromosomes in the pair. But as this is not ´dominant gene damageª, neither father nor mother show any sign of disease, they are healthy carriers of the gene. We are all carriers of about five recessive genes of this type, but as spouses are seldom carriers of exactly the same damaged gene(s), all will probably go well in the next generation.
- regulatory sequence - (regulatory region, regulatory area) is a segment of DNA where regulatory proteins such as transcription factors bind preferentially.
- ribosome - complex of rRNA and ribosomal proteins, bind mRNA and translate it into protein.
- RNA - RiboNucleic Acid. The intermediate nucleic acid involved in gene expression. It comes in 3 forms: tRNA, mRNA, rRNA.
- rRNA - ribosomal, translates mRNA into protein. rRNA provides the "scaffolding" on which many ribosomal proteins are assembled as 2 subunits that themselves assemble to form a ribosome. rRNA genes are localized to the nucleolus in the nucleus, a sometimes visible region of DNA usually constantly being transcribed.
- segmental aneuploidies - generated when a small piece of a chromosome is gained or lost during cell division, resulting in subchromosomal copy number (CN) changes.
- single umbilical artery - (SUA) Placental cord with only a single placental artery (normally paired). This abnormality can be detected by ultrasound (colour flow imaging of the fetal pelvis) and is used as an indicator for further prenatal diagnostic testing for chromosomal abnormalities and other systemic defects. (More? Prenatal Diagnosis | Ultrasound)
- telophase - Cell division term referring to the fifth mitotic stage, where the vesicles of the nuclear envelope reform around the daughter cells, the nucleoli reappear and the chromosomes unfold to allow gene expression to begin. This phase overlaps with cytokinesis, the division of the cell cytoplasm.
- telomere - regions at the end of chromosomes. Shortening of the telomeres is thought to be associated with cellular aging. The enzyme that maintains the telomere is called telomerase. Introducing this gene into a cell can extend the cells lifespan.
- topologically associating domain - (TAD) a self-interacting genomic region, DNA sequences within a TAD physically interact with each other more frequently than with sequences outside the TAD.
- transcription factor - a protein which binds to DNA activating (usually) gene expression. There are many different ways and forms that this activation can take place, but most transcription factors fall into specific classes (eg zinc fingers, helix loop helix).
- triple markers - alpha-fetoprotein, human chorionic gonadotropin, and unconjugated estriol. ))trisomy 21}}
- triploidy - genetic abnormality caused by one additional set of chromosomes, for a total of 69 chromosomes. Maternally present with albuminuria, edema, or hypertension. Extra maternally inherited chromosomes, microcephaly and an enlarged placenta that is enlarged and filled with cysts. Extra paternally inherited chromosomes, severe growth problems, enlarged head, and a small placenta. Non-mosaic triploidy is highly lethal, and is rarely observed in live births. ICD-11 LD42.0 Triploidy
- trisomy mosaicism - a rare chromosome disorder characterized by having an extra copy of a chromosome in a proportion, but not all, of a person’s cells.
- tRNA - transfer, binds single amino acids acts as a "donor' for protein synthesis.
- uniparental disomy - Genetic term referring to cells containing both copies of a homologous pair of chromosomes from one parent and none from the other parent.
|
|
|
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 27) Embryology Genetic risk maternal age. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Genetic_risk_maternal_age
- What Links Here?
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G