Molecular Development
Introduction
This page is a link to many different resources related to molecular development. I am 2.85 billion nucleotides of DNA, but so is a chimpanzee, and all this DNA encodes only about 20,000-25,000 protein-coding genes. In development, I am not that different from a mouse or a fly and many of the signals that regulate development are used time and time again.
We have come a long way from just observing development to now wanting to understand how the complex program of development is controlled. Using new research tools and some excellent animal models researchers have discovered common themes and mechanisms that tie all embryonic development together.
What is remarkable, given our biological diversity, is the strong evolutionary conservation of developmental mechanisms. This has been a boon in allowing the use of many (easier) model systems such as the genetist's tool the fruitfly, and the worm, frog, chicken, zebrafish and mouse (see other embryos page).
A continuing theme also seems to be the reuse of signals at different times and places within the embryo, for diiferent jobs. This has given rise to the concept of "switches" which by themselves may contain no "information" but to activate other genes or switches. Finally, you can imagine that of our 20,000-25,000 protein-coding genes, a large number of these may only be expressed during development or if reused, have a completely different role in the mature animal.
In terms of molecular mechanisms, the field of epigenetics has begun to florish with some recent important findings.
Molecular mechanisms of development is an exciting area and requires a variety of different skills. This page introduces only a few examples and should give you a feel for the topic. Note that each section of system notes has a page covering molecular development in that system. I have included information about the basic building blocks of proteins (amino acids).
| Factor Links: AMH | hCG | BMP | sonic hedgehog | bHLH | HOX | FGF | FOX | Hippo | LIM | Nanog | NGF | Nodal | Notch | PAX | retinoic acid | SIX | Slit2/Robo1 | SOX | TBX | TGF-beta | VEGF | WNT | Category:Molecular |
Starting Out
There are several different ways to begin to look at molecular development.
- look at the earliest molecular events involved in patterning the differentiation of cells- axis formation.
- or look at early events in patterning with differentiation and migration of cells to form the trilaminar embryo (gastrulation).
- or look at the signaling mechanisms and the different types of factors involved.
- or look at their role in the development of specific systems.
Signaling during development, though complex, can also be grouped into a few specific classes. These mechanisms have also been listed and described briefly on Signaling Mechanisms page.
For this section, Molecular Biology of the Cell (now indexed and available online from PubMed) and the extracted Cell Biology version are good reference texts. Also the key journals in this area are: Development, Genes and Development, PNAS, EMBOJ.
There is also detailed molecular information available from the abnormalities (page 2 of each section of notes) where there are links to the OMIM Database entry for specific genetic disorders.
Please understand that these notes are a new addition and are therefore under constant revision.
Gene Expression
Gene expression can often simply be described by the pathway: DNA -> mRNA -> Protein. While each cell contains the same genetic material, not all genes are ever expressed and the subset of genes expressed at any one time affects cellular differentiation and mature cell function. This can be impacted by both signaling and epigenetic mechanisms.
Protein
The individual amino acids that form all proteins can be represented by a standardised single letter code, or three letter code or by their entire name.
Single Letter Code
A - Alanine (Ala) | C - Cysteine (Cys) | D - Aspartic Acid (Asp) | E - Glutamic Acid (Glu) | F - Phenylalanine (Phe) | G - Glycine (Gly) | H - Histidine (His) | I - Isoleucine (Ile) | K - Lysine (Lys) |L - Leucine (Leu) | M - Methionine (Met) | N - Asparagine (Asn) | P - Proline (Pro) | Q - Glutamine (Gln) | R - Arginine (Arg) | S - Serine (Ser) | T - Threonine (Thr) | V - Valine (Val) | W - Tryptophan (Trp) | Y - Tyrosine (Tyr)
| Amino Acid | 3-Letter | 1-Letter | Side chain polarity | Side chain acidity or basicity of neutral species | Hydropathy index |
|---|---|---|---|---|---|
| Alanine | Ala | A | nonpolar | neutral | 1.8 |
| Arginine | Arg | R | polar | basic (strongly) | -4.5 |
| Asparagine | Asn | N | polar | neutral | -3.5 |
| Aspartic acid | Asp | D | polar | acidic | -3.5 |
| Cysteine | Cys | C | polar | neutral | 2.5 |
| Glutamic acid | Glu | E | polar | acidic | -3.5 |
| Glutamine | Gln | Q | polar | neutral | -3.5 |
| Glycine | Gly | G | nonpolar | neutral | -0.4 |
| Histidine | His | H | polar | basic (weakly) | -3.2 |
| Isoleucine | Ile | I | nonpolar | neutral | 4.5 |
| Leucine | Leu | L | nonpolar | neutral | 3.8 |
| Lysine | Lys | K | polar | basic | -3.9 |
| Methionine | Met | M | nonpolar | neutral | 1.9 |
| Phenylalanine | Phe | F | nonpolar | neutral | 2.8 |
| Proline | Pro | P | nonpolar | neutral | -1.6 |
| Serine | Ser | S | polar | neutral | -0.8 |
| Threonine | Thr | T | polar | neutral | -0.7 |
| Tryptophan | Trp | W | nonpolar | neutral | -0.9 |
| Tyrosine | Tyr | Y | polar | neutral | -1.3 |
| Valine | Val | V | nonpolar | neutral | 4.2 |
Hydropathy Index[2]
- a number representing the hydrophobic or hydrophilic properties of the amino acid sidechain.
- larger the number the more hydrophobic the amino acid.
- most hydrophobic amino acids are isoleucine (4.5) and valine (4.2), hydrophobic amino acids tend to be internal
- most hydrophilic amino acids are arginine (-4.5) and lysine (-3.9), hydrophilic amino acids are more commonly found towards the protein surface.
Class, amino acid, R group
Acidic
- D Asp Aspartic acid -CH2-COOH where both the O and the OH are bonded to the C
- E Glu Glutamic acid -CH2-CH2-COOH
Basic
- K Lys Lysine -CH2-CH2-CH2-CH2-NH3+
- R Arg Arginine -CH2-CH2-CH2-NH-C-(NH2)2 :where 2 NH2 groups are attached to the final C
- H His Histidine -CH2-<C-HN-HC=NH+-HC=> :where < chain > actually forms a pentagonal ring bonded at the C
Uncharged Polar
- S Ser Serine -CH2-OH
- T Thr Threonine -CH-CH3OH :where both the CH3 and the OH attach at the initial CH
- Y Tyr Tyrosine -CH2-<CH=CH-CH=COH-CH=CH-> :where < ... > forms a hexagonal ring with the OH at the far end
- N Asn Asparagine -CH2-CONH2 :where both the =O and the -NH2 attach to the C
- O Gln Glutamine -CH2-CH2-CONH2 :same as Asparagine but with an extra CH2
C Cys Cysteine -CH2-S-H
Non-Polar
- G Gly Glycine -H
- A Ala Alanine -CH3
- V Val Valine -CH-(CH3)2
- L Leu Leucine -CH2-CH-(CH3)2
- I Ile Isoleucine -CH-(CH3, CH2-CH3)
- P Pro Proline -<CH2-CH2-CH2> :where the other end of the ring attaches at the N by replacing an H
- F Phe Phenylalanine -CH2-<CH=CH-CH=CH-CH=CH-> :like tyrosine without the OH at the far end of the ring.
- M Met Methionine -CH2-CH2-S-CH3
- W Trp Tryptophan -CH2-<pentagonal ring with NH><Hexagonal Phenyl ring>
About Amino Acids
- A strong Disulfide Bridge (S-S) sometimes can be created between the sulfur in the side chains in two Cysteine amino acids. This plays a strong role in forming the 3 dimensional protein structure.
- Additional bonds occur between acidic and basic end groups of the side chains.
- Polar side groups like to remain in a polar environment such as the water surrounding the protein.
- A protein is most stable when in its lowest energy configuration.
Signaling Factors
| Factor Links: AMH | hCG | BMP | sonic hedgehog | bHLH | HOX | FGF | FOX | Hippo | LIM | Nanog | NGF | Nodal | Notch | PAX | retinoic acid | SIX | Slit2/Robo1 | SOX | TBX | TGF-beta | VEGF | WNT | Category:Molecular |
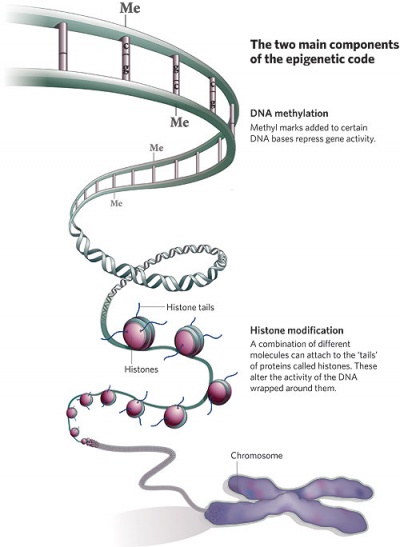
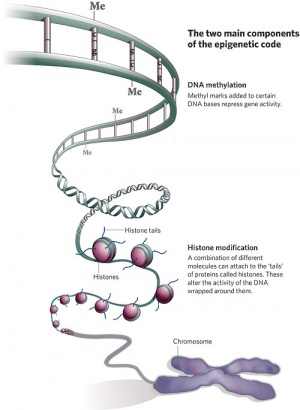
Epigenetics
References
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- Foldit - Amino Acids
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, May 6) Embryology Molecular Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Molecular_Development
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G