Developmental Signals - Wnt
| Embryology - 9 Mar 2026 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
A secreted glycoprotein patterning switch with different roles in different tissues and signaling has generally been divided into the canonical and non-canonical pathways. The name was derived from two drosophila phenotypes wingless and int and the gene was first defined as a protooncogene, int1.
Humans have 19 identified members, with the major subgroup of Wnts (WNT1, WNT3A, WNT8) signaling through activation of beta-catenin dependent transcription from at least 4 WNT genes encoding secreted glycoproteins. At least one wnt receptor called frizzled (FZD). Wnt7a is secreted protein and binds to extracellular matrix. The mechanism of Wnt distribution (free diffusion, restricted diffusion and active transport) and all its possible cell receptors are still being determined.
In the gastrointestinal system, Wnt maintains the pool of undifferentiated intestinal progenitor cells and control maturation and correct positioning of the Paneth cell (a differentiated intestinal cell type).
The Wnt/beta-catenin signal pathway activation has also been identified in several cancers and appears to drive cell-cycle progression.
If you are interested in this family of proteins, look also at Roel Nusse -The Wnt Homepage.
| Factor Links: AMH | hCG | BMP | sonic hedgehog | bHLH | HOX | FGF | FOX | Hippo | LIM | Nanog | NGF | Nodal | Notch | PAX | retinoic acid | SIX | Slit2/Robo1 | SOX | TBX | TGF-beta | VEGF | WNT | Category:Molecular |
Some Recent Findings
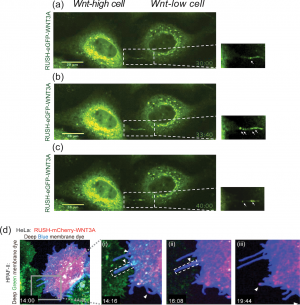
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References <pubmed limit=5>Wnt</pubmed> |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Human Wnt Family
| Table - Human Wnt Family | |||||
| Approved Symbol |
Approved Name | Previous Symbols |
Synonyms | Chromosome | |
|---|---|---|---|---|---|
| WNT1 | Wnt family member 1 | INT1 | 12q13.12 | ||
| WNT2 | Wnt family member 2 | INT1L1 | IRP | 7q31.2 | |
| WNT2B | Wnt family member 2B | WNT13 | XWNT2 | 1p13.2 | |
| WNT3 | Wnt family member 3 | INT4 | "MGC131950, MGC138321, MGC138323" | 17q21.31-q21.32 | |
| WNT3A | Wnt family member 3A | 1q42.13 | |||
| WNT4 | Wnt family member 4 | WNT-4 | 1p36.12 | ||
| WNT5A | Wnt family member 5A | hWNT5A | 3p14.3 | ||
| WNT5B | Wnt family member 5B | 12p13.33 | |||
| WNT6 | Wnt family member 6 | 2q35 | |||
| WNT7A | Wnt family member 7A | 3p25.1 | |||
| WNT7B | Wnt family member 7B | 22q13.31 | |||
| WNT8A | Wnt family member 8A | WNT8D | 5q31.2 | ||
| WNT8B | Wnt family member 8B | 10q24.31 | |||
| WNT9A | Wnt family member 9A | WNT14 | 1q42.13 | ||
| WNT9B | Wnt family member 9B | WNT15 | WNT14B | 17q21.32 | |
| WNT10A | Wnt family member 10A | 2q35 | |||
| WNT10B | Wnt family member 10B | "WNT-12, SHFM6" | 12q13.12 | ||
| WNT11 | Wnt family member 11 | 11q13.5 | |||
| WNT16 | Wnt family member 16 | 7q31.31 | |||
| Links: Developmental Signals - Wnt | OMIM Wnt1 | HGNC | Bmp Family | Fgf Family | Pax Family | R-spondin Family | Sox Family | Tbx Family | Wnt Family | |||||
| |||||
| Human WNT Family | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
WNT4
| Table - Human Wnt Family | ||||
| Approved Symbol |
Approved Name | Previous Symbols |
Synonyms | Chromosome |
|---|---|---|---|---|
| WNT4 | Wnt family member 4 | WNT-4 | 1p36.12 | |
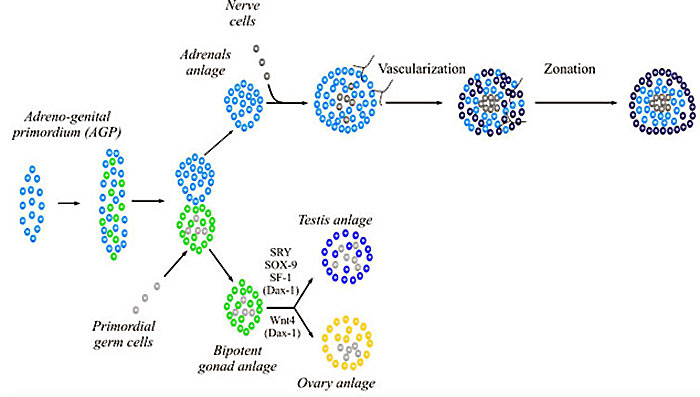
WNT4 adrenal and gonad early development[11]
- PubMed Search: WNT4
R-spondin
The 4 vertebrate secreted R-spondin proteins are agonists of the canonical Wnt/β-catenin signaling pathway (see reviews[12][13]).
R-spondin 1 (RSPO1) together with both WNT4 and FOXL2 required for ovary development.[14]).
| Table - Human R-spondin Family | ||||
| Approved Symbol |
Approved Name | Previous Symbols |
Synonyms | Chromosome |
|---|---|---|---|---|
| RSPO1 | R-spondin 1 | FLJ40906, RSPONDIN | 1p34.3 | |
| RSPO2 | R-spondin 2 | MGC35555 | 8q23.1 | |
| RSPO3 | R-spondin 3 | THSD2 | FLJ14440 | 6q22.33 |
| RSPO4 | R-spondin 4 | C20orf182 | dJ824F16.3 | 20p13 |
| Links: Developmental Signals - Wnt | Bmp Family | Fgf Family | Pax Family | R-spondin Family | Sox Family | Tbx Family | Wnt Family | ||||
| Human R-spondin Family | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||
- PubMed Search: R-spondin | HGNC - RSPO1
Function
Body Axis
Wnt signaling and the polarity of the primary body axis[16] "Data from diverse deuterostomes (frog, fish, mouse, and amphioxus) and from planarians (protostomes) suggest that Wnt signaling through beta-catenin controls posterior identity during body plan formation in most bilaterally symmetric animals."
Sensory
Hearing
Wnt9a Can Influence Cell Fates and Neural Connectivity across the Radial Axis of the Developing Cochlea.[17]
- "Vertebrate hearing organs manifest cellular asymmetries across the radial axis that underlie afferent versus efferent circuits between the inner ear and the brain. Therefore, understanding the molecular control of patterning across this axis has important functional implications. Radial axis patterning begins before the cells become postmitotic and is likely linked to the onset of asymmetric expression of secreted factors adjacent to the sensory primordium. This study explores one such asymmetrically expressed gene, Wnt9a, which becomes restricted to the neural edge of the avian auditory organ, the basilar papilla, by embryonic day 5 (E5). Radial patterning is disrupted when Wnt9a is overexpressed throughout the prosensory domain beginning on E3. Sexes were pooled for analysis and sex differences were not studied. Analysis of gene expression and afferent innervation on E6 suggests that ectopic Wnt9a expands the neural-side fate, possibly by re-specifying the abneural fate."
- Links: Inner Ear | Chicken Development
Vision
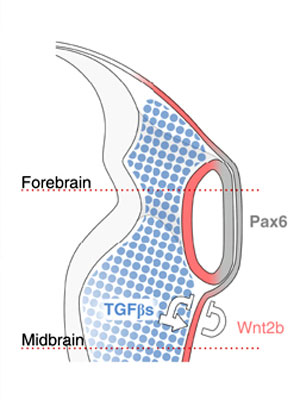
See figure from recent article, proposed embryological model summarizing how neural crest cells (NCCs) organize the eye: NCCs (blue) secrete TGF-βs, which signal to the non-lens ectoderm and dorsal optic vesicle. As a consequence, Wnt2b (red) is induced, and together they repress lens formation in the non-lens ectoderm. This leads to the alignment of Pax6 expression in the future lens and neural retina (grey).[18]
- Links: Lens
Neural
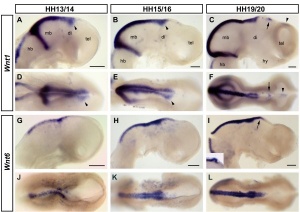
"Wnts are antagonised by Shh signalling. By demonstrating that Wnt4 expression in the thalamus is repressed by Shh from the ZLI we reveal an additional level of interaction between these two pathways and provide an example for the cross-regulation between patterning centres during forebrain regionalisation."[19]
Wnt receptor Frizzled-1 in presynaptic differentiation and function[20] "We examined the distribution of the Wnt receptor Frizzled-1 in cultured hippocampal neurons and determined that this receptor is located at synaptic contacts co-localizing with presynaptic proteins. Frizzled-1 was found in functional synapses detected with FM1-43 staining and in synaptic terminals from adult rat brain. Interestingly, overexpression of Frizzled-1 increased the number of clusters of Bassoon, a component of the active zone, while treatment with the extracellular cysteine-rich domain (CRD) of Frizzled-1 decreased Bassoon clustering, suggesting a role for this receptor in presynaptic differentiation. Consistent with this, treatment with the Frizzled-1 ligand Wnt-3a induced presynaptic protein clustering and increased functional presynaptic recycling sites, and these effects were prevented by co-treatment with the CRD of Frizzled-1."
- Links: Neural System Development)
Muscle
Wnt signaling promotes AChR aggregation at the neuromuscular synapse[21]
- "Wnt proteins regulate the formation of central synapses by stimulating synaptic assembly, but their role at the vertebrate neuromuscular junction (NMJ) is unclear. Wnt3 is expressed by lateral motoneurons of the spinal cord during the period of motoneuron-muscle innervation. ...Wnt3 does not signal through the canonical Wnt pathway to induce cluster formation. Instead, Wnt3 induces the rapid formation of unstable AChR micro-clusters through activation of Rac1, which aggregate into large clusters only in the presence of agrin."
Vascular
Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature[22] "The central nervous system (CNS) vasculature consists of a tightly sealed endothelium that forms the blood-brain barrier, whereas blood vessels of other organs are more porous. Wnt7a and Wnt7b encode two Wnt ligands produced by the neuroepithelium of the developing CNS coincident with vascular invasion. Using genetic mouse models, we found that these ligands directly target the vascular endothelium and that the CNS uses the canonical Wnt signaling pathway to promote formation and CNS-specific differentiation of the organ's vasculature"
Respiratory
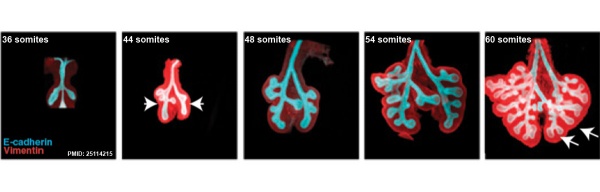
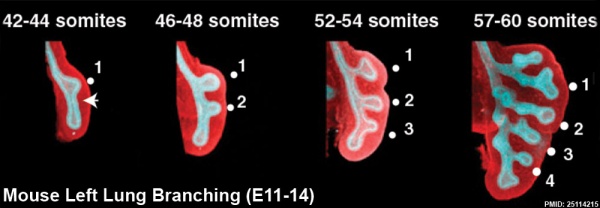
This study used knockout mice to show the role of Wnt signalling in bronchial branching morphogenesis between E10 to E14.[23] The images below are from a normal branching sequence with a mesenchyme (red) and epithelium (blue) marker. Knocking out Frizzled 2 receptor impacted on both tube shape and branch-point formation in the developing lung.
- Links: respiratory | mouse
Pancreas
inhibition of Wnt signaling, in both organoid and human stem cell cultures, promotes insulin-producing cell generation.[24]
- Links: pancreas
Signaling Pathway
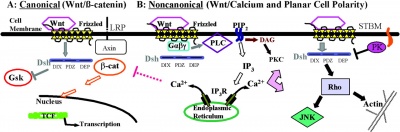
At least 3 separate signaling pathways have been identified in relation to Wnt singnaling.
- Canonical pathway
- Non-canonical or planar cell polarity pathway
- Wnt-Calcium ion pathway
Canonical Pathway
- Wnt binds the surface receptor Frizzled (Fz) and LRP5/6 receptor complex
- Induces the stabilization of beta-catenin (through the DIX and PDZ domains of Dishevelled and other factors including Axin, glycogen synthase kinase 3 and casein kinase 1)
- Beta-catenin translocates into the nucleus
- Beta-catenin complexes with members of the LEF/TCF family of transcription factors.
- Transcriptional induction of target genes.
- Beta-catenin is then exported from the nucleus and degraded via the proteosomal machinery.
Example - early development of the sinoatrial node cells in the heart.
Non-Canonical Pathway
(Planar Cell Polarity Pathway)
- Wnt binds the surface receptor Frizzled (Fz) independent of the LRP5/6 receptor complex.
- Acts through Dishevelled (PDZ and DEP domains)
- Mediates cytoskeletal (microfilament) changes through activation of the small GTPases Rho and Rac.
Wnt-Calcium Ion Pathway
- Wnt binds the surface receptor Frizzled (Fz) and mediates activation of heterotrimeric G-proteins.
- Acts internally through Dsh, phospholipase C, calcium-calmodulin kinase 2 (CamK2) and protein kinase C (PKC).
- has various cellular functions including cell adhesion and motility.
Protein Data
- FUNCTION: PROBABLE DEVELOPMENTAL PROTEIN. SIGNALING BY WNT-7A ALLOWS SEXUALLY DIMORPHIC DEVELOPMENT OF THE MULLERIAN DUCTS (BY SIMILARITY).
- SUBCELLULAR LOCATION: POSSIBLY SECRETED AND ASSOCIATES WITH THE EXTRACELLULAR MATRIX.
- TISSUE SPECIFICITY: EXPRESSION IS RESTRICTED TO PLACENTA, KIDNEY, TESTIS, UTERUS, FETAL LUNG, AND FETAL AND ADULT BRAIN.
- SIMILARITY: BELONGS TO THE WNT FAMILY.
- FUNCTION: PROBABLE DEVELOPMENTAL PROTEIN. SIGNALING BY WNT-7A ALLOWS SEXUALLY DIMORPHIC DEVELOPMENT OF THE MULLERIAN DUCTS.
- SUBCELLULAR LOCATION: POSSIBLY SECRETED AND ASSOCIATES WITH THE EXTRACELLULAR MATRIX.
- DISEASE: MALE MICE LACKING WNT-7A FAIL TO UNDERGO REGRESSION OF THE MULLERIAN DUCT AS A RESULT OF THE ABSENCE OF THE RECEPTOR FOR MULLERIAN-INHIBITING SUBSTANCE. WNT7A-DEFICIENT FEMALES ARE INFERTILE BECAUSE OF ABNORMAL DEVELOPMENT OF THE OVIDUCT AND UTERUS, BOTH OF WHICH ARE MULLERIAN DUCT DERIVATIVES.
- SIMILARITY: BELONGS TO THE WNT FAMILY.
(data from Expasy)
OMIM
About OMIM "Online Mendelian Inheritance in Man OMIM is a comprehensive, authoritative, and timely compendium of human genes and genetic phenotypes. The full-text, referenced overviews in OMIM contain information on all known mendelian disorders and over 12,000 genes. OMIM focuses on the relationship between phenotype and genotype. It is updated daily, and the entries contain copious links to other genetics resources." OMIM
References
- ↑ Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ & Yang Y. (2003). Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J. Cell Biol. , 162, 899-908. PMID: 12952940 DOI.
- ↑ 2.0 2.1 Moti N, Yu J, Boncompain G, Perez F & Virshup DM. (2019). Wnt traffic from endoplasmic reticulum to filopodia. PLoS ONE , 14, e0212711. PMID: 30794657 DOI.
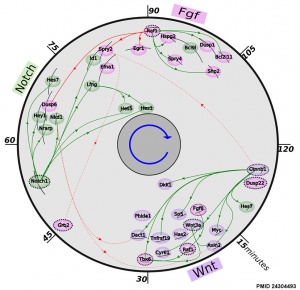
- ↑ 3.0 3.1 Fongang B & Kudlicki A. (2013). The precise timeline of transcriptional regulation reveals causation in mouse somitogenesis network. BMC Dev. Biol. , 13, 42. PMID: 24304493 DOI.
- ↑ Sahebdel F, Parvaneh Tafreshi A, Arefian E, Roussa E, Nadri S & Zeynali B. (2022). Wnt/β-catenin signaling pathway is involved in early dopaminergic differentiation of trabecular meshwork-derived mesenchymal stem cells. J Cell Biochem , 123, 1120-1129. PMID: 35533251 DOI.
- ↑ Hernández-Martínez R, Ramkumar N & Anderson KV. (2019). p120-catenin regulates WNT signaling and EMT in the mouse embryo. Proc. Natl. Acad. Sci. U.S.A. , 116, 16872-16881. PMID: 31371508 DOI.
- ↑ Grainger S & Willert K. (2018). Mechanisms of Wnt signaling and control. Wiley Interdiscip Rev Syst Biol Med , , e1422. PMID: 29600540 DOI.
- ↑ Baranowska Körberg I, Hofmeister W, Markljung E, Cao J, Nilsson D, Ludwig M, Draaken M, Holmdahl G, Barker G, Reutter H, Vukojević V, Clementson Kockum C, Lundin J, Lindstrand A & Nordenskjöld A. (2015). WNT3 involvement in human bladder exstrophy and cloaca development in zebrafish. Hum. Mol. Genet. , 24, 5069-78. PMID: 26105184 DOI.
- ↑ Roarty K, Shore AN, Creighton CJ & Rosen JM. (2015). Ror2 regulates branching, differentiation, and actin-cytoskeletal dynamics within the mammary epithelium. J. Cell Biol. , 208, 351-66. PMID: 25624393 DOI.
- ↑ Mattes B, Weber S, Peres J, Chen Q, Davidson G, Houart C & Scholpp S. (2012). Wnt3 and Wnt3a are required for induction of the mid-diencephalic organizer in the caudal forebrain. Neural Dev , 7, 12. PMID: 22475147 DOI.
- ↑ Carmon KS & Loose DS. (2010). Development of a bioassay for detection of Wnt-binding affinities for individual frizzled receptors. Anal. Biochem. , 401, 288-94. PMID: 20227380 DOI.
- ↑ Val P, Lefrançois-Martinez AM, Veyssière G & Martinez A. (2003). SF-1 a key player in the development and differentiation of steroidogenic tissues. Nucl. Recept. , 1, 8. PMID: 14594453 DOI.
- ↑ de Lau WB, Snel B & Clevers HC. (2012). The R-spondin protein family. Genome Biol. , 13, 242. PMID: 22439850 DOI.
- ↑ Jin YR & Yoon JK. (2012). The R-spondin family of proteins: emerging regulators of WNT signaling. Int. J. Biochem. Cell Biol. , 44, 2278-87. PMID: 22982762 DOI.
- ↑ Biason-Lauber A. (2012). WNT4, RSPO1, and FOXL2 in sex development. Semin. Reprod. Med. , 30, 387-95. PMID: 23044875 DOI.
- ↑ Ferrer-Vaquer A, Piliszek A, Tian G, Aho RJ, Dufort D & Hadjantonakis AK. (2010). A sensitive and bright single-cell resolution live imaging reporter of Wnt/ß-catenin signaling in the mouse. BMC Dev. Biol. , 10, 121. PMID: 21176145 DOI.
- ↑ Petersen CP & Reddien PW. (2009). Wnt signaling and the polarity of the primary body axis. Cell , 139, 1056-68. PMID: 20005801 DOI.
- ↑ Munnamalai V, Sienknecht UJ, Duncan RK, Scott MK, Thawani A, Fantetti KN, Atallah NM, Biesemeier DJ, Song KH, Luethy K, Traub E & Fekete DM. (2017). Wnt9a Can Influence Cell Fates and Neural Connectivity across the Radial Axis of the Developing Cochlea. J. Neurosci. , 37, 8975-8988. PMID: 28821654 DOI.
- ↑ 18.0 18.1 Grocott T, Johnson S, Bailey AP & Streit A. (2011). Neural crest cells organize the eye via TGF-β and canonical Wnt signalling. Nat Commun , 2, 265. PMID: 21468017 DOI.
- ↑ 19.0 19.1 Quinlan R, Graf M, Mason I, Lumsden A & Kiecker C. (2009). Complex and dynamic patterns of Wnt pathway gene expression in the developing chick forebrain. Neural Dev , 4, 35. PMID: 19732418 DOI.
- ↑ Varela-Nallar L, Grabowski CP, Alfaro IE, Alvarez AR & Inestrosa NC. (2009). Role of the Wnt receptor Frizzled-1 in presynaptic differentiation and function. Neural Dev , 4, 41. PMID: 19883499 DOI.
- ↑ Henriquez JP, Webb A, Bence M, Bildsoe H, Sahores M, Hughes SM & Salinas PC. (2008). Wnt signaling promotes AChR aggregation at the neuromuscular synapse in collaboration with agrin. Proc. Natl. Acad. Sci. U.S.A. , 105, 18812-7. PMID: 19020093 DOI.
- ↑ Stenman JM, Rajagopal J, Carroll TJ, Ishibashi M, McMahon J & McMahon AP. (2008). Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science , 322, 1247-50. PMID: 19023080 DOI.
- ↑ Kadzik RS, Cohen ED, Morley MP, Stewart KM, Lu MM & Morrisey EE. (2014). Wnt ligand/Frizzled 2 receptor signaling regulates tube shape and branch-point formation in the lung through control of epithelial cell shape. Proc. Natl. Acad. Sci. U.S.A. , 111, 12444-9. PMID: 25114215 DOI.
- ↑ Yung T, Poon F, Liang M, Coquenlorge S, McGaugh EC, Hui CC, Wilson MD, Nostro MC & Kim TH. (2019). Sufu- and Spop-mediated downregulation of Hedgehog signaling promotes beta cell differentiation through organ-specific niche signals. Nat Commun , 10, 4647. PMID: 31604927 DOI.
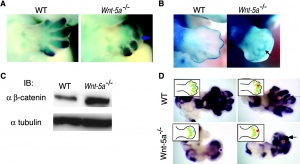
- ↑ Westfall TA, Brimeyer R, Twedt J, Gladon J, Olberding A, Furutani-Seiki M & Slusarski DC. (2003). Wnt-5/pipetail functions in vertebrate axis formation as a negative regulator of Wnt/beta-catenin activity. J. Cell Biol. , 162, 889-98. PMID: 12952939 DOI.
Reviews
de Jaime-Soguero A, Abreu de Oliveira WA & Lluis F. (2018). The Pleiotropic Effects of the Canonical Wnt Pathway in Early Development and Pluripotency. Genes (Basel) , 9, . PMID: 29443926 DOI.
Inestrosa NC & Arenas E. (2010). Emerging roles of Wnts in the adult nervous system. Nat. Rev. Neurosci. , 11, 77-86. PMID: 20010950 DOI.
Petersen CP & Reddien PW. (2009). Wnt signaling and the polarity of the primary body axis. Cell , 139, 1056-68. PMID: 20005801 DOI.
Franco CA, Liebner S & Gerhardt H. (2009). Vascular morphogenesis: a Wnt for every vessel?. Curr. Opin. Genet. Dev. , 19, 476-83. PMID: 19864126 DOI.
van Amerongen R & Nusse R. (2009). Towards an integrated view of Wnt signaling in development. Development , 136, 3205-14. PMID: 19736321 DOI.
Search PubMed: Wnt
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- The Wnt gene Homepage by Roel Nusse
- Atlas of Genetics and Cytogenetics in Oncology and Haematology - The WNT Signaling Pathway and Its Role in Human Solid Tumors
- Mouse Wnt genes
- Comparative table of all vertebrate Wnt genes
- alignments of all mammalian Wnt genes
- Other Wnt pages
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2026, March 9) Embryology Developmental Signals - Wnt. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Developmental_Signals_-_Wnt
- © Dr Mark Hill 2026, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G