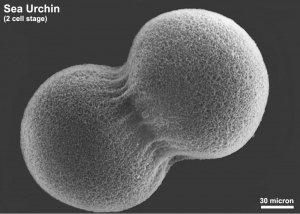
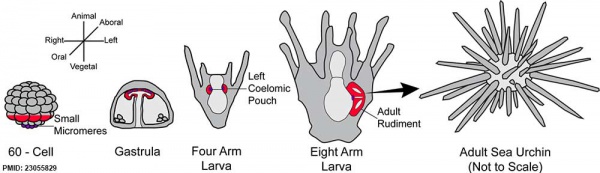
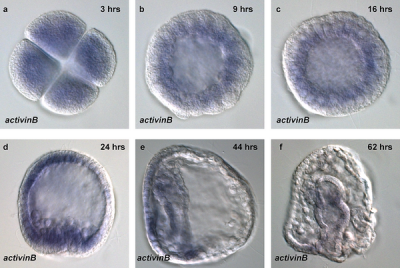
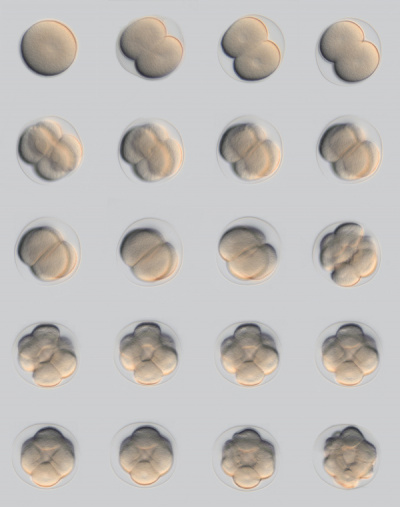
Sea Urchin Development
| Embryology - 1 Mar 2026 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
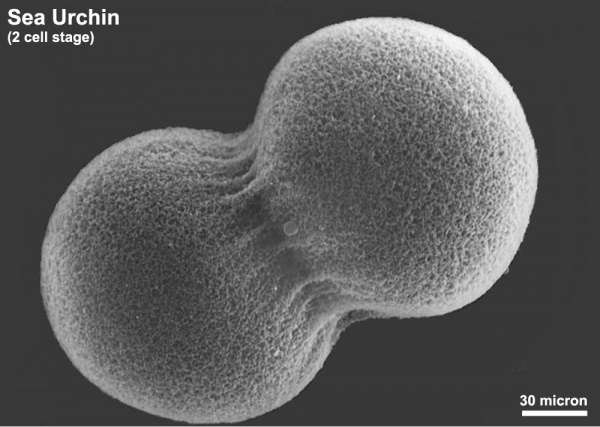
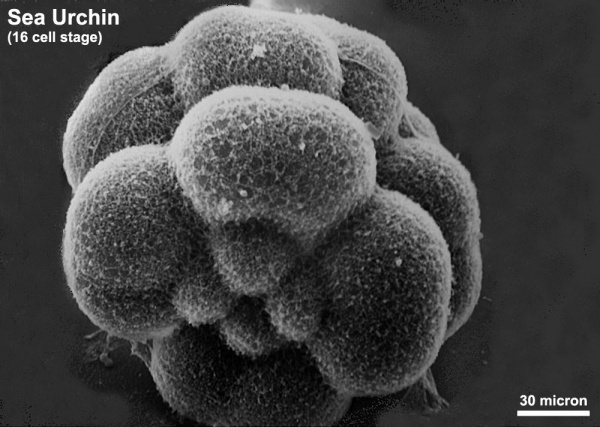
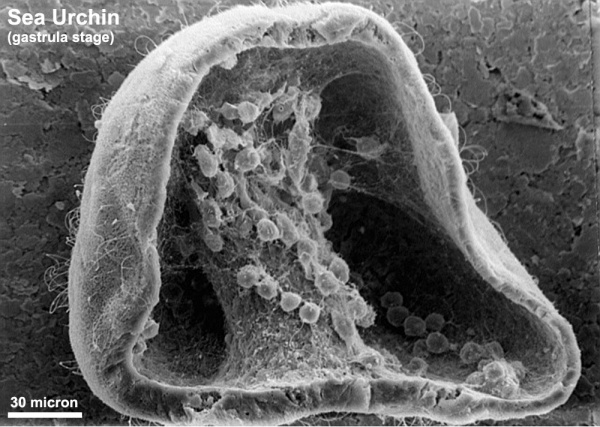
The sea urchin embryo initially undergoes ten cycles of cell division forming a single epithelial layer enveloping a blastocoel, followed by gastrulation producing the three germ layers. Historically, one of the earliest systems used in developmental biology (H. Driesch, 1892; J. Loeb, 1893; Lillie, 1912[1] Spermatozoa Chemotaxis; S. Horstadius, 1928), this embryo system has been used recently to study early molecular controls of patterning and axis formation.
- Links: Category:Sea Urchin
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Sea Urchin Embryology |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Movies
| <html5media height="300" width="280">File:Spermatozoa chemotaxis PMID23183693.mp4</html5media> | Spermatozoa Chemotaxis
|
- Links: Hans Driesch | Movies
Images
Sea Urchin- activin B expression
Sea Urchin- early embryo cleavage pattern
Sea urchin ectoderm patterning model
Early Development
Endomesoderm Induction
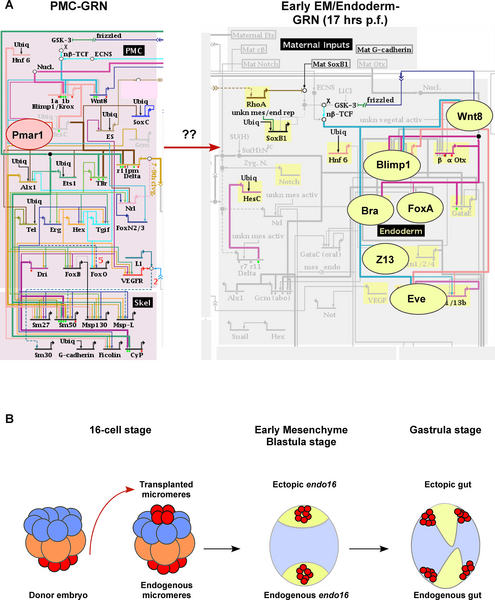
Sea Urchin-endomesoderm induction[8] Figure illustrates gene regulatory networks required for the early developmental process of endomesoderm induction.
Ectoderm Development
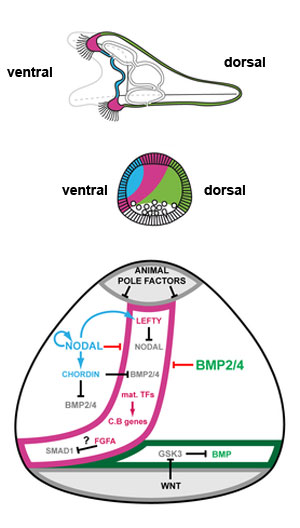
Changes in identity of ectodermal territories following perturbations of Nodal or BMP signaling and novel model of ectoderm patterning[9]
Schemes describing the morphology of control embryos and perturbed embryos.
(A) control embryo. The thick ciliated epithelium of the ciliary band is restricted to a belt of cells at the interface between the ventral and dorsal ectoderm.
(B) Nodal morphant. Most of the ectoderm differentiates into an expanded large ciliary band. An animal pole domain is nevertheless present in these embryos as shown by the presence of the apical tuft and at the molecular level by the expression of apical domain marker genes. In these embryos, the ectoderm surrounding the blastopore differentiates into dorsal ectoderm.
(C) embryo overexpressing Nodal. Most of the ectoderm differentiates into ventral ectoderm. A ciliary band-like ectoderm forms at the animal pole and in the ectoderm surrounding the blastopore.
(D) BMP2/4 morphants. An ectopic ciliary band forms in the dorsal ectoderm in addition to the normal ciliary band.
(E) bmp2/4 overexpressing embryo. All the ectoderm has a dorsal identity. The animal pole domain is largely absent. The triradiated stars represent the spicule rudiments.
(F) Proposed model for regionalization of the ectoderm of the sea urchin embryo through restriction of the ciliary band fate by Nodal and BMP signaling. Maternal factors such as SoxB1 promote the early expression of ciliary band genes within the ectoderm. Nodal signaling on the ventral side promotes differentiation of the ventral ectoderm and stomodeum and represses the ciliary band fate probably through the activity of Goosecoid as well as of additional repressors. Nodal induces its antagonist Lefty, which diffuses away from the ventral ectoderm up to the presumptive ciliary band territory. Within the ventral ectoderm, Nodal induces expression of bmp2/4 and of its antagonist chordin. Chordin prevents BMP signaling within the ventral ectoderm and probably within the presumptive ciliary band region. At blastula stages, protein complexes containing BMP2/4 and Chordin can diffuse towards the dorsal side to specify dorsal fates. In the dorsal ectoderm, BMP signaling strongly repress the ciliary band fate partly by inducing the expression of the irxA repressor. A high level of MAP kinase activity resulting from FGFA signaling in the lateral ectoderm likely contributes to maintain a low level of Nodal and BMP signaling within the presumptive ciliary band region by phosphorylating Smad1/5/8 and Smad2/3 in the linker region, which inhibits their activity. The presence of Chordin and Lefty in the prospective ciliary band allows expression of ciliary band genes to be maintained in this region. The ectoderm surrounding the blastopore differentiates into dorsal ectoderm likely because it receives Wnt signals that antagonize GSK3 and promote BMP signaling.
(G) In the absence of Nodal signaling, both the ventral and the dorsal inducing signals are not produced, ciliary band genes are not repressed and unrestricted MAP kinase signaling promotes differentiation of the ventral and dorsal ectoderm into neural ectoderm and ciliary band. The genes or proteins that are inactive are represented in light grey.
Wnt patterning
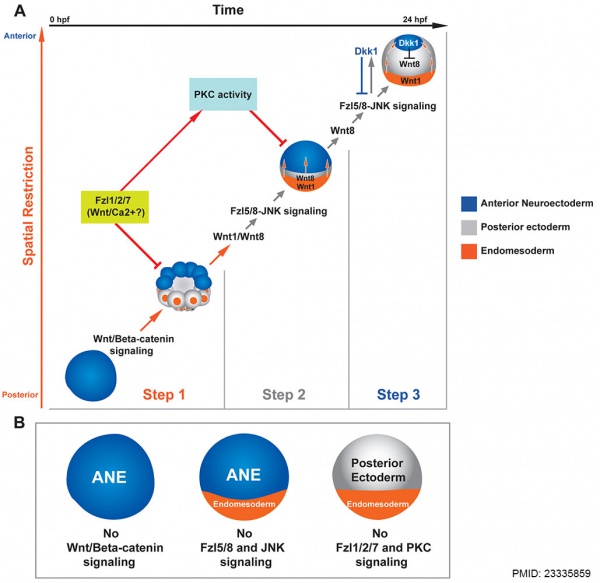
Sea urchin Wnt patterning model[10]
History
| Embryology History - Hans Driesch (1867 – 1941) was a German experimental embryology. From 1891 to 1901 he worked in Naples at the Marine Biological Station. His experimental work was designed to establish a formulation for development and ended by adopting an Aristotlean teleological theory of entelechy. | 
Hans Driesch (1867 – 1941) |
| Embryology History - Hans Driesch manipulating blastomeres of sea urchin eggs. | 
|
Historic Images
| Historic Disclaimer - information about historic embryology pages |
|---|
| Pages where the terms "Historic" (textbooks, papers, people, recommendations) appear on this site, and sections within pages where this disclaimer appears, indicate that the content and scientific understanding are specific to the time of publication. This means that while some scientific descriptions are still accurate, the terminology and interpretation of the developmental mechanisms reflect the understanding at the time of original publication and those of the preceding periods, these terms, interpretations and recommendations may not reflect our current scientific understanding. (More? Embryology History | Historic Embryology Papers) |
Bailey FR. and Miller AM. Text-Book of Embryology (1921) New York: William Wood and Co.
References
- ↑ Lillie FR. (1912). THE PRODUCTION OF SPERM ISO-AGGLUTININS BY OVA. Science , 36, 527-30. PMID: 17735765 DOI.
- ↑ Uchida A & Yajima M. (2018). An optogenetic approach to control protein localization during embryogenesis of the sea urchin. Dev. Biol. , 441, 19-30. PMID: 29958898 DOI.
- ↑ Shevidi S, Uchida A, Schudrowitz N, Wessel GM & Yajima M. (2017). Single nucleotide editing without DNA cleavage using CRISPR/Cas9-deaminase in the sea urchin embryo. Dev. Dyn. , 246, 1036-1046. PMID: 28857338 DOI.
- ↑ Yajima M, Umeda R, Fuchikami T, Kataoka M, Sakamoto N, Yamamoto T & Akasaka K. (2010). Implication of HpEts in gene regulatory networks responsible for specification of sea urchin skeletogenic primary mesenchyme cells. Zool. Sci. , 27, 638-46. PMID: 20695779 DOI.
- ↑ Croce JC & McClay DR. (2010). Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development , 137, 83-91. PMID: 20023163 DOI.
- ↑ Stamateris RE, Rafiq K & Ettensohn CA. (2010). The expression and distribution of Wnt and Wnt receptor mRNAs during early sea urchin development. Gene Expr. Patterns , 10, 60-4. PMID: 19853669 DOI.
- ↑ Kaupp UB. (2012). 100 years of sperm chemotaxis. J. Gen. Physiol. , 140, 583-6. PMID: 23183693 DOI.
- ↑ Sethi AJ, Angerer RC & Angerer LM. (2009). Gene regulatory network interactions in sea urchin endomesoderm induction. PLoS Biol. , 7, e1000029. PMID: 19192949 DOI.
- ↑ Saudemont A, Haillot E, Mekpoh F, Bessodes N, Quirin M, Lapraz F, Duboc V, Röttinger E, Range R, Oisel A, Besnardeau L, Wincker P & Lepage T. (2010). Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet. , 6, e1001259. PMID: 21203442 DOI.
- ↑ Range RC, Angerer RC & Angerer LM. (2013). Integration of canonical and noncanonical Wnt signaling pathways patterns the neuroectoderm along the anterior-posterior axis of sea urchin embryos. PLoS Biol. , 11, e1001467. PMID: 23335859 DOI.
Reviews
Kominami T & Takata H. (2004). Gastrulation in the sea urchin embryo: a model system for analyzing the morphogenesis of a monolayered epithelium. Dev. Growth Differ. , 46, 309-26. PMID: 15367199 DOI.
Articles
Search Pubmed
Search Pubmed: Sea Urchin Development
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- Developmental Biology - The Early Development of Sea Urchins
- 4D Map of the Virtual Sea Urchin Embryo A research project aimed at creating a graphical interface that provides a spatial and temporal context in which to browse the genomic regulatory network of the sea urchin during the early stages of its embryonic development.
- Sea urchin embryology lab This document was prepared for a school-teachers' workshop.
- Stanford University - Sea Urchins
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2026, March 1) Embryology Sea Urchin Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Sea_Urchin_Development
- © Dr Mark Hill 2026, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G