Sea Squirt Development: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 4: | Line 4: | ||
Sea squirts (ascidians, Ascidiacea) are filter feeding marine animals, primitive chordates, that exist in many shapes and sizes. They reproduce by either sexual or asexual (budding) and develop through a laval (tadpole) to an adult phase. Some species have transparent eggs and embryos simplifying developmental imaging. | Sea squirts (ascidians, Ascidiacea) are filter feeding marine animals, primitive chordates, that exist in many shapes and sizes. They reproduce by either sexual or asexual (budding) and develop through a laval (tadpole) to an adult phase. Some species have transparent eggs and embryos simplifying developmental imaging. | ||
See also the historic paper identifying cell lineages in the sea squirt embryo. | |||
{{Ref-Conklin1905}} | |||
{{Animals}} | {{Animals}} | ||
| Line 19: | Line 19: | ||
|} | |} | ||
{| class="wikitable mw-collapsible mw-collapsed" | |||
! More recent papers | |||
|- | |||
| [[File:Mark_Hill.jpg|90px|left]] {{Most_Recent_Refs}} | |||
Search term: [http://www.ncbi.nlm.nih.gov/pubmed/?term=Sea+Squirt+Embryology ''Sea Squirt Embryology''] | |||
<pubmed limit=5>Sea Squirt Embryology</pubmed> | |||
|} | |||
==Early Development== | ==Early Development== | ||
Latest revision as of 15:46, 22 October 2016
Introduction
Sea squirts (ascidians, Ascidiacea) are filter feeding marine animals, primitive chordates, that exist in many shapes and sizes. They reproduce by either sexual or asexual (budding) and develop through a laval (tadpole) to an adult phase. Some species have transparent eggs and embryos simplifying developmental imaging.
See also the historic paper identifying cell lineages in the sea squirt embryo.
Conklin EG. The Organization and Cell-Lineage of the Ascidian Egg (1905) J. Acad., Nat. Sci. Phila. 13, 1.
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Sea Squirt Embryology <pubmed limit=5>Sea Squirt Embryology</pubmed> |
Early Development
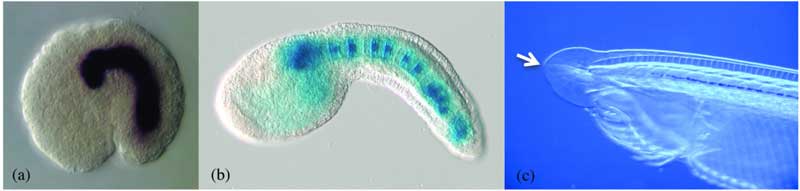
- (b) Ciona intestinalis notochord[4]
Neural
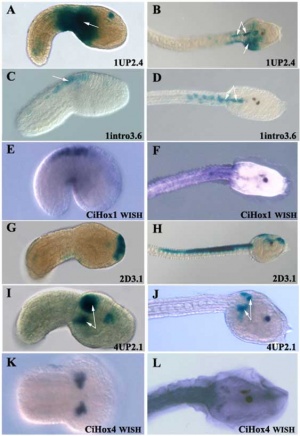
- ascidians lack a segmented hindbrain, but have restricted expression patterns of anterior Hox genes.[5]
Historic
Manual of Human Embryology by Franz Keibel and Franklin P. Mall (1910).
- "Conklin (1895) has been able to determine in ascidian eggs, even before cleavage begins, the existence of organ-forming substances, one of which, the myoplasm, that has to do with the formation of muscle tissue, is clearly recognizable and can be followed through successive stages of development into formed muscle."
References
Reviews
<pubmed>21558365</pubmed>
Articles
Search Pubmed
Search Pubmed: Sea Squirt Development
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- Ascidian Home Page for United States Ascidian News - Published twice a year since 1975.
- The Dutch Ascidians Homepage
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 26) Embryology Sea Squirt Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Sea_Squirt_Development
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G