Fly Development
| Embryology - 27 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
This page introduces the fly, drosophila, as a developmental model organism. The small drosophila fruitfly has been used by genetisists for many years now and much is now understood about its development in relation to gene expression and regulatory mechanisms.
In recent years, using developmental mutants, many mechanisms of development in the fly have been shown to be almost identical to those seen in humans and other animals. In fact, these developmental mechanisms have become the "paradigm" for our understanding of development.
The fruitfly (drosophila) was and is the traditional geneticist's tool. It has been transformed to an magnificent tool for the embryologist, with many developmental mechanisms being uncovered in this system combined with homolgy gene searches in other species.
Drosophila researchers have received to date received 5 Nobel prizes (1933, 1995, 2011). The most recent in 2011 "for their discoveries concerning the activation of innate immunity".
There is also a difference in basic body structure between males and females, males lack the seventh abdominal segment (A7) present in females. This has recently been shown to be due to a down-regulation of epidermal growth factor receptor (EGFR) activity and fewer histoblasts in the male A7 in the early pupae.[1]
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Drosophila Development |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Taxon
melanogaster group
Taxonomy Id: 32346 Rank: species group
Genetic code: Translation table 1 (Standard) Mitochondrial genetic code: Translation table 5 Lineage( abbreviated ): Eukaryota; Metazoa; Arthropoda; Tracheata; Hexapoda; Insecta; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; Ephydroidea; Drosophilidae; Drosophila
Development
The drosophila lifespan varies with temperature and is about 30 days at 29 °C.
A series of papers published between 1976 to 1979 by Turner and Mahowald, characterised the stages of drosophila development in beautiful scanning electron microscope (SEM) images.[11][12][13]
Hox Genes
|

|
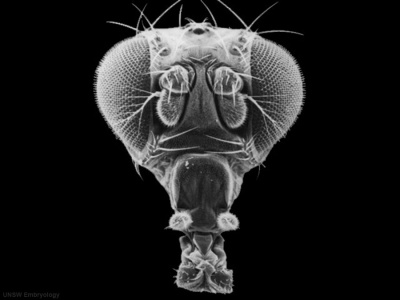
| Fly wild-type head[14] | Fly antennapedia mutant head[14] |
This is the classic mutation that gave rise to the discovery of Hox genes and other genes related to body pattern formation. In this mutant during development the fly embryo incorrectly positioned where (antenna) should have be two legs (pedia)[14]. The discovery of this mutant in Walter Gehring's lab opened up the field of developmental genes and this field has been rewarded with the 1995 Nobel prize in Medicine.
- Links: Hox | 1995 Nobel Prize
Hippo Genes
The Hippo (Hpo) pathway, first identified in Drosophila, controls organ size by regulating cell proliferation (inhibition) and apoptosis (induction). In contrast, the TOR signalling pathway regulates organ size by stimulating cell growth, thus increasing cell size.
| Fly Phenotype (dorsal view head thorax SEM) | |
|---|---|
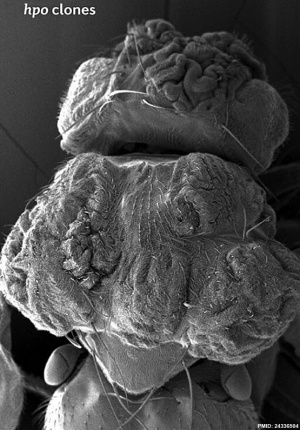
|

|
| Hippo-type (hpo) | Wild-type (WT) |
| Image source[15] |
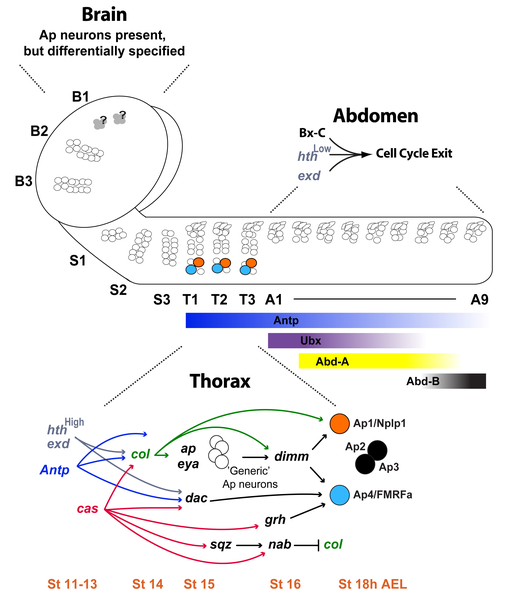
Neural Development
Summary of neural development from neural stem cell population and the gene regulation involved.[9]
References
- ↑ Foronda D, Martín P & Sánchez-Herrero E. (2012). Drosophila Hox and sex-determination genes control segment elimination through EGFR and extramacrochetae activity. PLoS Genet. , 8, e1002874. PMID: 22912593 DOI.
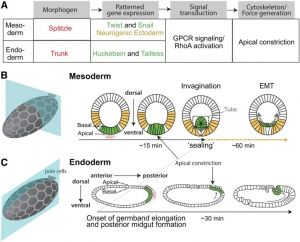
- ↑ 2.0 2.1 Martin AC. (2020). The Physical Mechanisms of Drosophila Gastrulation: Mesoderm and Endoderm Invagination. Genetics , 214, 543-560. PMID: 32132154 DOI.
- ↑ Lammers NC, Galstyan V, Reimer A, Medin SA, Wiggins CH & Garcia HG. (2020). Multimodal transcriptional control of pattern formation in embryonic development. Proc. Natl. Acad. Sci. U.S.A. , 117, 836-847. PMID: 31882445 DOI.
- ↑ Trush O, Liu C, Han X, Nakai Y, Takayama R, Murakawa H, Carrillo JA, Takechi H, Hakeda-Suzuki S, Suzuki T & Sato M. (2019). N-cadherin orchestrates self-organization of neurons within a columnar unit in the Drosophila medulla. J. Neurosci. , , . PMID: 31175213 DOI.
- ↑ Yang Y, Zhou M, Fang Q & Shen HB. (2019). AnnoFly: Annotating Drosophila Embryonic Images Based on an Attention-Enhanced RNN Model. Bioinformatics , , . PMID: 30601935 DOI.
- ↑ Zhu JY, Fu Y, Nettleton M, Richman A & Han Z. (2017). High throughput in vivo functional validation of candidate congenital heart disease genes inDrosophila. Elife , 6, . PMID: 28084990 DOI.
- ↑ Hartenstein V, Younossi-Hartenstein A, Lovick JK, Kong A, Omoto JJ, Ngo KT & Viktorin G. (2015). Lineage-associated tracts defining the anatomy of the Drosophila first instar larval brain. Dev. Biol. , 406, 14-39. PMID: 26141956 DOI.
- ↑ de Navascués J & Modolell J. (2010). The pronotum LIM-HD gene tailup is both a positive and a negative regulator of the proneural genes achaete and scute of Drosophila. Mech. Dev. , 127, 393-406. PMID: 20580820 DOI.
- ↑ 9.0 9.1 Karlsson D, Baumgardt M & Thor S. (2010). Segment-specific neuronal subtype specification by the integration of anteroposterior and temporal cues. PLoS Biol. , 8, e1000368. PMID: 20485487 DOI.
- ↑ Yu L, Lee T, Lin N & Wolf MJ. (2010). Affecting Rhomboid-3 function causes a dilated heart in adult Drosophila. PLoS Genet. , 6, e1000969. PMID: 20523889 DOI.
- ↑ Turner FR & Mahowald AP. (1976). Scanning electron microscopy of Drosophila embryogenesis. 1. The structure of the egg envelopes and the formation of the cellular blastoderm. Dev. Biol. , 50, 95-108. PMID: 817949
- ↑ Turner FR & Mahowald AP. (1977). Scanning electron microscopy of Drosophila melanogaster embryogenesis. II. Gastrulation and segmentation. Dev. Biol. , 57, 403-16. PMID: 406152
- ↑ Alper PR. (1975). Letter: Lawsuit motivation. J Leg Med (N Y) , 3, 7. PMID: 1081572
- ↑ 14.0 14.1 14.2 Turner FR & Mahowald AP. (1979). Scanning electron microscopy of Drosophila melanogaster embryogenesis. III. Formation of the head and caudal segments. Dev. Biol. , 68, 96-109. PMID: 108157
- ↑ Johnson R & Halder G. (2014). The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov , 13, 63-79. PMID: 24336504 DOI.
Journals
Developmental Dynamics
Journal of Neurobiology
- Special Issue: Unexpected Roles for Morphogens in the Development and Regeneration of the CNS Volume 64, Issue 4 (15 September 2005)
- Marques G. Morphogens and synaptogenesis in Drosophila. J Neurobiol. 2005 Sep 15;64(4):417-34.
Online Textbooks
Molecular Biology of the Cell (4th Edn) Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter. New York: Garland Publishing; 2002.
- Figure 21-24. Synopsis of Drosophila development from egg to adult fly
- Drosophila Begins Its Development as a Syncytium
- Figure 21-2. Homologous proteins functioning interchangeably in the development of mice and flies
Developmental Biology (6th Edn) Gilbert, Scott F. Sunderland (MA): Sinauer Associates, Inc.; c2000.
- Early Drosophila Development
- Snapshot Summary: Drosophila Development and Axis Specification
- Limb formation
Search NLM Online Textbooks- "drosophila development" : Molecular Biology of the Cell | Molecular Cell Biology | The Cell- A molecular Approach
Reviews
Sen A & Cox RT. (2017). Fly Models of Human Diseases: Drosophila as a Model for Understanding Human Mitochondrial Mutations and Disease. Curr. Top. Dev. Biol. , 121, 1-27. PMID: 28057297 DOI.
Tadros W & Lipshitz HD. (2005). Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. , 232, 593-608. PMID: 15704150 DOI.
Pereanu W & Hartenstein V. (2004). Digital three-dimensional models of Drosophila development. Curr. Opin. Genet. Dev. , 14, 382-91. PMID: 15261654 DOI.
Voas MG & Rebay I. (2004). Signal integration during development: insights from the Drosophila eye. Dev. Dyn. , 229, 162-75. PMID: 14699588 DOI.
Weigmann K, Klapper R, Strasser T, Rickert C, Technau G, Jäckle H, Janning W & Klämbt C. (2003). FlyMove--a new way to look at development of Drosophila. Trends Genet. , 19, 310-1. PMID: 12801722 DOI.
Articles
Shingleton AW, Das J, Vinicius L & Stern DL. (2005). The temporal requirements for insulin signaling during development in Drosophila. PLoS Biol. , 3, e289. PMID: 16086608 DOI.
Search PubMed
Search Aug2005 "drosophila development" 13228 reference articles of which 1899 were reviews.
Search Pubmed: fly development | drosophila development
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- Genetics Society of America Annual Drosophila Conference
Databases
There are a number of excellent internet resources for Fly development.
- Flybase - A Database of the Drosophila Genome http://flybase.bio.indiana.edu/
- The Interactive Fly - looks at genes and development http://www.sdbonline.org/fly/aimain/1aahome.htm This site is very well organized and allows an exploration of the molecular mechanisms of development. Remember that this is where molecular mammalian embryology all started through homology.
- Flybrain - An Online Atlas and Database of the Drosophila Nervous System http://flybrain.neurobio.arizona.edu/
- FlyServer - A Drosophila Image Database and A Drosophila Multimedia Database http://pbio07.uni-muenster.de/
- NCBI Taxonomy Browser | Drosophila Genome Resources
- AnnoFly - Annotating Drosophila Embryonic Images Based on an Attention-Enhanced RNN Model
Fly Pages
- Development of Drosophila - by Katherine Plewes, Becky Wong and Leon W. Browder http://www.ucalgary.ca/UofC/eduweb/virtualembryo/flies.html
- BIO 114 Virtual Fly - lntroductory Biology Lab course at the WKU Glasgow extended campus http://bioweb.wku.edu/courses/Biol114/Vfly1.asp
- Chapter 13A: Drosophila Development - Kenyon College http://biology.kenyon.edu/courses/biol114/Chap13/Chapter_13A.html
- Journal of Visualized Experiments Live Imaging Of Drosophila melanogaster Embryonic Hemocyte Migrations
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 27) Embryology Fly Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Fly_Development
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G