User:Z5020317: Difference between revisions
No edit summary |
No edit summary |
||
| Line 347: | Line 347: | ||
<pubmed> 18671204 </pubmed> | <pubmed> 18671204 </pubmed> | ||
==L8 assessment== | |||
==History== | |||
Prenatal Genetic Diagnosis was historically conducted post-implantation in the first 2 trimesters <ref> Blackburn, S.L. (2003) '''Maternal, Fetal & Neonatal physiology: a Clinical perspective''' (2nd ed.). Seattle: Saudners </ref>, testing for overall fetal growth, complications of pregnancy, birth defects as well as chromosomal or genetic abnormalities<ref> Sadler T.W.(2012) '''Langman's Medical Embryology''' (12th ed.) Philadelphia: Lipincott, Wiliams & Wilkins, a Wolters Kluwer Business </ref>. Due to advancements in Artificial Reproductive Technologies ( ART) in the past 2 decades , these genetic tests are now conducted as a part of Preimplantation Genetic Diagnosis (PGD) <ref><pubmed>20638568</pubmed></ref> and art used to eliminate the rsik of passing on a genetic disorder to an offspring, before implantation, it ha now become a preventative measure. . In 1990 Preimplantation genetic testing was first used in 1990 on humans to diagnose recessive X-linked diseases in the embryos of pregnant women undergoing IVF. <ref><pubmed>2330030</pubmed></ref> PGD is offered as an alternative to prenatal diagnosis in detecting genetic disorders in high risk couples <ref><pubmed>26168107</pubmed></ref> and avoid passing an inherited disease to the offspring, by selecting genetically "normal" embryos for implantation ,to give the best chance of successful implantation , stable pregnancies and healthy children. | |||
<ref><pubmed>23499002</pubmed></ref> | |||
==Genetic Techniques== | |||
===PCR=== | |||
====Description==== | |||
Polymerase chain reaction (PCR) amplifies DNA specific to genetic sequence of interest . PCR was developed by Kay Mullis in the 1980s, for which he was awarded the Nobel prize for chemistry in 1993. <ref> Pubmed Docs (2015) Polymerase Chain Reaction (PCR) Pubmen. Retrieved from [http://www.ncbi.nlm.nih.gov/probe/docs/techpcr/]</ref> This technique enables clinicians to monitor and diagnose diseases using minute samples such as embryonic cells, blood & tissue. <ref>Roche (2015) PCR: How We Copy DNA. Roche Molecular Systems Inc. retrieved From [http://molecular.roche.com/pcr/Pages/Process.aspx]</ref> PCR has many important applications like DNA fingerprinting, genetic mapping, detection of viruses and bacteria as well as being used to detect genetic disorders, as a part of PGD, in conjunction with IVF. It is used to detect molecular abnormalities such as single gene disorders like Tay Sach, Cycstic fibrosis, Duchenne Muscular Dystrophy, Thalassemia, Huntington disease, Spinal muscular atrophy and many more. Molecular and Genetic analysis require a significant amount of DNA, which we would not have without PCR. It revolutionized the study of DNA, replacing all previous recombinant DNA technology. <ref> Mullins, K., Francois, F.& Gibbs R.A. (1994 ) The Polymerase Chain Reaction. p3. Springer- Science +Business Media. Birkhauser, Boston | |||
Available at [https://books.google.com.au/books?hl=en&lr=&id=gjrTBwAAQBAJ&oi=fnd&pg=PR5&dq=kary+mullis+PCR&ots=mpzAyRh5ZY&sig=PQ4goNoKJ90tb3-MkPk62zrTGbw#v=onepage&q=kary%20mullis%20PCR&f=false] </ref> | |||
====Procedure==== | |||
Sample are obtained from the the blastocyst, a polar body biopsy or the blastomeres stages of the embryo. | |||
Stage 1 Denaturing: separating the target strands of DNA | |||
The obtained sample is heated to roughly 90 degrees celcius, this heat causes breaks the relatively weak bonds between nucleotides that form DNA. the double stranded DNA to split into 2 single strands of DNA that are used as templates. | |||
Stage 2 Annealing: Binding the Primers to the target DNA sequence | |||
PCR will only copy the target sequence of DNA specified by specific PCR primer. These synthesized primers Oligonucleotides are small artificial pieces of DNA. | |||
TAQ polymerase enzyme synthesize 2 new strands of DNA duplicate to the single sample DNA stand template indicated by these primers. During this stage the reaction is cooled to a temperature between 40-60 degrees Celsius. | |||
Stage 3- Extension- making copies | |||
Each of these 2 copies are then used again as templates generating 2 further replications This cycle can occur as many 30 -40 times within a couple hours leading to billions of extra copies of the original DNA segment. The optimal temperature for the further replication is roughly 72 degrees Celsius. | |||
{| | |||
|- | |||
| '''PCR Cycle''' | |||
| '''Target Copies''' | |||
|- | |||
| 1 | |||
|2 | |||
|- | |||
| 2 | |||
|4 | |||
|- | |||
| 3 | |||
| 8 | |||
|- | |||
| 4 | |||
| 16 | |||
|- | |||
| 5 | |||
|32 | |||
|- | |||
|6 | |||
|64 | |||
|- | |||
| 7 | |||
|128 | |||
|- | |||
| 8 | |||
|256 | |||
|- | |||
| 9 | |||
|512 | |||
|- | |||
| 10 | |||
|1024 | |||
|- | |||
| 15 | |||
| 32,768 | |||
|- | |||
| 20 | |||
|1,048,578 | |||
|- | |||
| 25 | |||
|33,554,432 | |||
|- | |||
| 30 | |||
|1,073,741,842 | |||
|-. | |||
|} | |||
This process mediated by a thermocycler machine that is programmed to alter the temperature of the reaction every couple of minutes, perpetuating the cycle of DNA denaturing and synthesis. | |||
This process, generates exponential exact copy of the original template DNA sequence. | |||
====Advantages & Disadvantages==== | |||
ADVANTAGES | |||
*fast and inexpensive way of copying a target sequence of DNA | |||
*aids the diagnosis of single cell defects | |||
*rapid generation of results within a couple hours | |||
*can start from a single molecule of DNA, therefore is more sensitive | |||
*has a flexible primer design enabling | |||
*overcame previous issues associated with the limited availability of sites of end | |||
* generates the DNA copies exponentially | |||
*enables DNA amplification required for molecular and genetic analysis | |||
*Valid diagnostic method in selecting unaffected embryos for embryo transfer <ref> Dreesen, J., Drusedaul, M., Smeets, H., Die-Smulders, C., Coonen, E., Dumoulin, J., Gielen, M., Evers, J., Herbergers, J. & Geradets, J. (2008) Validation of preimplantation genetic diagnosis by PCR analysis: genotype comparison of the blastomere and corresponding embryo, implications for clinical practice. Mol. Hum. Reprod. 14 (10):573-579.doi: 10.1093/molehr/gan052. Retrieved from [http://molehr.oxfordjournals.org/content/14/10/573.short] </ref> <ref><pubmed>20966460</pubmed></ref> | |||
DISADVANTAGES | |||
*only during this exponential DNA replication phase can we determine the starting quantity sequence contained in the original sample ( template DNA strand) | |||
*PCR reaction is limited by the presence of inhibitors present in the sample : | |||
reagent inhibitors | |||
*self annealing due to the accumulation of the product -stopping exponential amplification for the target sequence and reach a plateau | |||
quantification of the end point of reaction of PCR products unreliable - therefore that is why we require real time quantitative RT-PCR necessary | |||
<ref> NIS (2015) Polymerase Chain Reaction (PCR) National Human Genome Research Institute retrieved from [https://www.genome.gov/10000207]</ref> | |||
===Fluorescent In Situ Hybridisation (FISH)=== | |||
====Description==== | |||
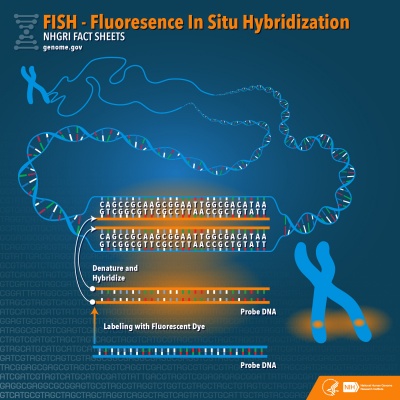
Fluorescence in situ hybridization (FISH) the one of the most effective and rapid <ref><pubmed>26338801</pubmed></ref> Preimplantation Genetic Diagnosis (PGD). This techniques used to locate a specific DNA sequences within a chromosome. FISH facilitates the clinical diagnosis of chromosomal abnormalities indicated by sequential duplications, deletions and rearrangements of chromosome, that are usually missed with microscopic analysis. This technique is especially relevant for female embryos with X-linked diseases,<ref><pubmed>20809319</pubmed></ref> that have no other mutation specific tests [http://www.ncbi.nlm.nih.gov/pubmed/20809319] As part of PGD, samples are collected from various stages of the embyro,and they are able to conducted tests on the blastocyst, a polar body biopsy and the blastomeres <ref><pubmed>21748341</pubmed></ref>. FISH is 99% effective when used in conjunction with competitive Genomic Hybridisation (CGH) to diagnose chromosomal abnormalities.<ref><pubmed>26338801</pubmed></ref> | |||
====Procedure==== | |||
Samples are collected from either the blastocyst, a polar body biopsy or the blastomeres stages of the embryo <ref><pubmed>21748341</pubmed></ref>. | |||
DNA strands are heated and denatured causing their the individual DNA strands to break apart | |||
Probes are single complimentary stands of DNA a that have been tagged with small chemical agents that glow brightly in the presence of a specific region on a chromosome. These specific probes then hybridize and join to their complementary DNA strand. | |||
The fluorescent tags enable the researchers to correctly identify the presence or lack thereof and location of the specific chromosomes that they are testing for. <ref><pubmed>17876073</pubmed></ref> | |||
Researcher analyse the results by identifying the number and the relative location of the fluorescent dots generated by the FISH images.<ref><pubmed>17970921</pubmed></ref> | |||
The probe will not fully hybridise if there has been a duplication or a deletion of the DNA - indicating chromosomal and sex chromosomal anomalies like trisomies and aneuploidies. <ref>NIS (2015) Flourescent in Situ Hydridization (FISH) National Human Genome Research Institute retrieved from [https://www.genome.gov/10000206]</ref> | |||
Different probes are used for different purposes: <ref>Unique, Rare Chromosome Disorder Support Group (2013) Fluorescence in situ hybridisation (FISH) Rarechromo.org retrieved from [http://www.rarechromo.org/information/Other/FISH%20FTNW.pdf]</ref> | |||
*'''Locus specific tags''' detect very small imbalances, and locate isolated small portions | |||
of genes within a chromosome | |||
*'''Alphoid/Centomeric Repeat probes''' are developed from the repetitive sequence located in the middle region of each chromosome, useful in determining the number of chromosomes , detecting rearrangement and when used in conjunction with locus probes to determine the absence of genetic material on a chromosome. | |||
*'''Paint Probes''' are collections of smaller probes with their own stains that bind to a different section on the chromosome - allowing the full chromosome too be labeled a unique colour, this "full colour map" can be used to know the spectral karyotype- full chromosomal mapping is useful in examining chromosomal abnormalities. | |||
[[File:Fluorescent In Situ Hybridisation (FISH).jpg|400px]] | |||
====Advantages & Disadvantages==== | |||
ADVANTAGES | |||
*FISH is 99% effective when used in conjunction with Competitive Genomic Hybridisation (CGH) to diagnose chromosomal abnormalities.<ref><pubmed>26338801</pubmed></ref> | |||
*rapid generation of results | |||
*enables the number and size of specific chromosomes to be known | |||
*identifies very small translocations and aneuploidies that occur within chromosomes, that would usually be missed under microscopic analysis | |||
*indicates whether the accessed chromosomes are normal <ref>Iyer, B. (2013) SlideShare PreImplantation genetic diagnosis(pgd) Retrieved Oct 2, 2015 [http://www.slideshare.net/iyerbk/pre-implantation-genetic-diagnosis-pgd?related=1]</ref> | |||
*tests for the most common chromosomal abnormalities - down syndrome chromosomes 13,16,18,21 and 22 <ref><pubmed>21749752</pubmed></ref> | |||
*can be used to identify and remove inheritable X-linked disease or sex chromasome anomalies such as Duchennes Muscular Dystrophy, hemophilia, ectodermal dysplasia <ref><pubmed>17876073</pubmed></ref> | |||
* it may be used to compare the chromosomal gene arrangements in related species | |||
DISADVANTAGES | |||
*FISH destroys all cells tested <ref> O'Connor, C. (2008) Fluorescence in situ hybridization (FISH). Nature Education 1(1):171. retrieved from [http://www.nature.com/scitable/topicpage/fluorescence-in-situ-hybridization-fish-327] </ref> | |||
*it does not fully access all chromosomes ( only ~ 12), | |||
*there is a 40 % chance that chromosomal aneuploidy is occuring and not being targeted by this test <ref>Iyer, B. (2013) SlideShare PreImplantation genetic diagnosis(pgd) Retrieved Oct 2, 2015 [http://www.slideshare.net/iyerbk/pre-implantation-genetic-diagnosis-pgd?related=1]</ref> | |||
* analysis of results is dependent upon the dot quality impacted by hybridisation efficiency or the camera sensitivity <ref><pubmed>17970921</pubmed></ref> | |||
*some studies indicate that using FISH on a day-3 embryo biopsy decreases the rate of live births <ref><pubmed>26168107</pubmed></ref> | |||
PMID 11325751 | |||
===Array Comparative Genomic Hybridisation (aCGH)=== | |||
====Description==== | |||
aCGH also known as Microarray analysis efficiently scans the entire genome for chromosomal imbalances. CGH was initially developed to detect the number of changes in a solid tumor mass. It uses 2 genomes comparing the sample to the control, with each labeled in a different fluorescent dye<ref><pubmed>1876176</pubmed></ref>. Earlier CGH techniques were limited by the resolution of the imaging <ref> Lichter, P., et al. Comparative genomic hybridization: Uses and limitations. Seminars in Hematology 37, 348–357 (2000)</ref> , these initial limitations were overcome by using Microarrays in conjunction with CGh to improve the resolution of the imaging, Array Comparative Genomic Hybridisation (aCGH). This method compares sample and control microarrayed slides containing small segments of DNA (probes). <ref> Lucito, R., et al. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Research 13, 2291–2305 (2003) </ref> the Probes used will vary according from the small (25-85 base pairs) oligonucleotides manufactured to highlight different target sequences, to the very large genomic clones (80,000- 200,00 base pairs), and as these are significantly smaller than the traditional metaphase chromosomes used for CGH, generating a higher resolution of image. <ref><pubmed>22467166</pubmed></ref>.aCGH is used as a diagnostic tool for prenatal detection f chromosomal abnormalities <ref><pubmed>19012303</pubmed></ref>. | |||
====Procedure==== | |||
<ref> Theisen, A. (2008) Microarray-based Comparative Genomic Hybridization (aCGH). Nature Education 1(1):45. Retrieved from [http://www.nature.com/scitable/topicpage/microarray-based-comparative-genomic-hybridization-acgh-45432] </ref> | |||
*sample is obtained (skin, blood or fetal cells) and DNA is obtained. | |||
As a part of PGD fetal cell samples are collected from; the fertilized egg polar bodies, the blastomere (day 3 embryo) or the blastocyst/tropoectoderm stage (day 5 embryo). | |||
*Sample DNA is labeled with one fluorescent dye, and the control DNA is labeled with a different colored fluorescent dye. the control DNA is used as the base point of reference. | |||
heated and denatured single DNA strands then hybridize to their complementary single strand probes | |||
*these are then combined and applied to a microarray | |||
* the results are run through a computer program and a digital imaging system is used to quantify the results( fluorescent intensities of the labeled probes). | |||
simple explanation of aCGH : [Genomics Education (2014) Genetic Testing for Health: aCGH|https://vimeo.com/84757281] | |||
<ref>Genomics Education (2014) Genetic Testing for Health: aCGH [video file] Vimeo [https://vimeo.com/84757281]</ref> | |||
The fluorescent ratio and the hybridization signal at different locations on the genome of the control DNA are used to identify any variances present in the sample DNA,. aCGH facilitates the clinical diagnosis of submicroscopic chromosomal duplication, deletion and rearrangements indicative of chromosomal disorders such as trisomies 1-22and specific sex linked disorders. | |||
Duplications in the DNA are displayed by the computer program as spikes/ peaks over an established threshold and deletions in DNA are displayed by the computer program as spikes/ toughs beneath this threshold | |||
-computer screening image- | |||
-drawn diagram of method- | |||
====Advantages & Disadvantages==== | |||
''ADVANTAGES:'' | |||
*multiple applications: prenatal genetic diagnosis, cancer diagnosis, <ref><pubmed>20193845</pubmed></ref> genetic screening for developmental delay (learning disabilities) <ref><pubmed>17309648</pubmed></ref>. or congenital anomalies that are suspected to be genetic in origin <ref>NHS Array Comparitive Genomic Hybridisation (arrayCGH) pgh foundation . retrieved from [http://www.phgfoundation.org/file/5237/]</ref> | |||
*trace represents all chromosomes present in the human genome chromosomes 1-22 and the X & Y chromosomes, and therefore is the most accurate method for testing whole embryo aneuploidy | |||
*aCGH has been extensively tested, and Validated, and is now used world wide. | |||
*detects deletions and additions and rearrangements as well as amplification, of the WHOLE genome, simultaneously. | |||
*detects submicroscopic alterations | |||
*used as a diagnostic tool for prenatal detection of chromosomal abnormalities <ref><pubmed>22034057</pubmed></ref>. | |||
*provides high resolution genomewide screening of segmental genomic copy number variations | |||
*very accurate when used in conjunction with FISH | |||
*Alone is more reliable and in detecting chromosomal abnormalities, with a higher implantation success rate, than FISH <ref><pubmed>20494259</pubmed></ref>. | |||
*Allows an in depth research focus upon specific types of rearrangements within selected chromosomal regions, a recent particular area of interest is subtelomeric and pericentromeric rearrangements | |||
*reduces the risk of failed implantation and miscarriage, improving the chance of a healthy baby <ref>Iyer, B. (2013) SlideShare PreImplantation genetic diagnosis(pgd) Retrieved Oct 2, 2015 [http://www.slideshare.net/iyerbk/pre-implantation-genetic-diagnosis-pgd?related=1]</ref> | |||
''DISADVANTAGES:'' | |||
*translocations and inversions of DNA are not detected | |||
*limited ability diagnosing specific polyploidies such as triploidy <ref> Unique, Rare Chromosome Disorder Support Group (2013) Microarray-based Comparative Genomic Hybridiation (array CGH) Rarechromo.org retrieved from [http://www.rarechromo.org/information/other/array%20cgh%20ftnw.pdf] </ref> | |||
*will not detect mosaicism detection <20% (cultures where >1 of 5 cells are trisomy 12) | |||
*will not detect balanced chromosomal rearrangement | |||
*will not detect duplications or deletions <80kb | |||
*will not detect point mutations within genes <ref>Washington department of education (CGH- FAQ for physicians. Signature Genomic LAboratories, LLC. Retrieved from [https://depts.washington.edu/dbpeds/Lab%20Tests/SignatureCGH-physician_FAQ.pdf]</ref> | |||
* will not detect chromosomal position of genomic gains | |||
*will not detect loos of heterozygosity (LOH) or Absence of heterozygosity (AOH) <ref>WiCell Research Institute Inc. (2012) Comparative Genomic Hybridization Microarray. | |||
retrieved from [http://www.wicell.org/home/cytogenetic-services/cgh-microarray/array-comparative-genomic-hybridization-acgh.cmsx]</ref> | |||
===Next Generation Sequencing=== | |||
====Description==== | |||
The development and advances within ART in the past 20 years, as well as the increasing popularity of IV, has lead to an influx of new technologies developed to screen embryos for chromosomal anomalies, which are covered by the umbrella term of Next generation Sequencing (NGS). NGS is a general term used to describe all of the new and emerging screening techniques currently being introduced and used as part of PGD for IVF . NGS screens for single gene disorders as well as conducting extensive and very comprehensive chromosome diagnosis by sequencing, counting, and accurately assembling millions of DNA reads, simultaneously. <ref><pubmed>23499002</pubmed></ref> There is a movementfor NGS to replace the other limited testing techniques and be used as the standard. | |||
====Procedure==== | |||
Samples are collected from either blastomere or the blastocyst/tropoectoderm and processed for analysis by a computer system. | |||
The methodology of each process is unique to the technique being used. | |||
====Advantages & Disadvantages==== | |||
''ADVANTAGES:'' | |||
*cheaper | |||
*opens new diagnostic possibilities | |||
*accurately tests all 24 chromosomes | |||
*tests for the presence of monogenic diseases of known genetic background | |||
*reduces the number of biopsies required for diagnosis | |||
*higher detection rate of small translocations | |||
*highly accurate is testing for compound point mutations, chromosomal duplication, deletions and insertions <ref><pubmed>23312231</pubmed></ref> | |||
*it accurately detects chromosomal aneuploidy and unbalanced rearrangement <ref><pubmed>25685330</pubmed></ref> | |||
*NGS single gene disorder screenings can conducted in conjunction with PCR comprehensive chromosomal screening. | |||
*Reduces human error | |||
*better detects the presence of mosaicism | |||
*work well in conjunction with CGH and aCGH as part of PGD, improving the chances of IVF. | |||
<ref> Morris, R. S. (2015 ) Next generation sequencing for PCG|PGS|CCS. IVF1 retrieved from [http://www.ivf1.com/next-generation-sequencing-for-pgd] </ref> | |||
''DISADVANTAGES:'' | |||
there is limited information available to clinical applications of NGS <ref><pubmed>26100406</pubmed></ref> | |||
==Lab Attendance== | ==Lab Attendance== | ||
--[[User:Z5020317|Z5020317]] ([[User talk:Z5020317|talk]]) 13:46, 7 August 2015 (AEST) | --[[User:Z5020317|Z5020317]] ([[User talk:Z5020317|talk]]) 13:46, 7 August 2015 (AEST) | ||
| Line 362: | Line 571: | ||
--[[User:Z5020317|Z5020317]] ([[User talk:Z5020317|talk]]) 12:43, 25 September 2015 (AEST) | --[[User:Z5020317|Z5020317]] ([[User talk:Z5020317|talk]]) 12:43, 25 September 2015 (AEST) | ||
--[[User:Z5020317|Z5020317]] ([[User talk:Z5020317|talk]]) 11:45, 9 October 2015 (AEDT) | |||
Revision as of 10:45, 9 October 2015
Lab 1 Assessment
Your Lab assessment now requires you to find a 2 recent research references on fertilisation or in vitro fertilisation.
Paste each reference on your page, as shown in the class.
Write below each reference a brief summary of the research article methods and findings.
The summary for each need not be more than 3-4 paragraphs in length.
This will need to be completed before next weeks laboratory.
The use of r-hFSH in treatment of idiopathic male factor infertility before ICSI.
look at this [1]
PMID 26166637
The objective of this investigations was to evaluate whether or not the pre-treatment use of Recombinant Human Follicle Stimulating Hormone (r-hFSH), could be used to treat common male infertility issues as Idiopathic Oligozoospermia, to improve their clinical results of Intracytoplasmic Sperm Injection (ICSI).
They selected a sample of 82 couples that were infertile due to the Idiopathic male factor, from the Biofertility Center in Rome, Italy between May 2013- April 2014 and randomly selected a treatment group of 36 males, Group A, and control group of 46 men, Group. The men in treatment Group A were subcutaneously injected with 150 IU of R-hFSH; Gonad F© (recombinant Human FSH) 3 times per week, for a 3 month period. The subjects in the control Group B were injected with a placebo and therefore received no treatment. After the three month period all 82 couples underwent a cycle of ICSI. In each group fertilisation, rate of implantation, rate of clinical pregnancy and the rate of miscarriage were assessed.
They found that the rate fertilisation was comparable in both treatment groups, although treatment group A had a significantly higher implantation rate and clinical pregnancy rate as well as a statistically significant lower rate of miscarriage (P<0.05). The conclusion that they drew was that the treatment use of r-hFSH to treat Idiopathic males infertility prior to ICSI can improve the clinical rate of pregnancy, and increase the rate of implantation as well as decrease the chances of early pregnancy loss.
Effect of hepatitis C virus infection on the outcomes of in vitro fertilization.
look at this [2]
PMID 26131230
The objective of this experiment was to investigate the effects that the Hepatitis C Virus (HCV) infection has upon the outcome In Vitro Fertilisation (IVF). From January 2008 - December 2013 a large sample of data was collected from couples opting for IVF at the Centre for Reproductive Medicine, General Hospital of Tianjin Medical University. These 1424 couples were tested for the HCV antibody (HCV-Ab) and were then separated into 3 groups. Group A contained 90 couples where the female tested HCV positive, Group B had 78 couples in which the male partner tested HCV positive and Group C - was the control group, of 1256 couples that both tested negative for HCV. The control group was also matched in age and in accordance with the IVF ovarian hyper stimulation protocol. These couples then underwent IVF and the results were closely monitored.
Initially the sperm was collected and its concentration, volume, motility, normal sperm morphology percentage (NMP) and Teratozoospermia index (TZI) were assessed, and again tested for the presence of HCV-Ab. The women were tested with Follicle Stimulating Hormone (FSH) on days 2-4 of their Menstrual cycle, within the 6 months of IVF treatment. They underwent Ovarian Hyperstilumation, and intramuscular injections of chronic Gonadotropin to mature the Oocytes. Transvaginal collection of mature Oocytes occurred roughly 36 hours later. 3 days later 2 high quality embryos were then transferred back into the uterus and oral progesterone was administered to support the Luteal phase of menstruation cycle. 14 days after the embryo transfer, pregnancy can be detected by testing for increasing levels of hCG in the blood, and can later, be clinically diagnosed through the presence of gastrointestinal sac during an abdominal ultrasound.
They found that all the groups (HVC positive male, HCV positive female , HCV negative couples) had similar results in terms of age, ovarian response, endometrial thickness, duration of Gonadotropin administration, embryo transfer and day of hCG administration, and no obvious differences within the sperm parameters; motility, concentration , PR differences, volume and NMP, TZI. They proceeded to investigate the effects of HCV on pregnancy and found no notable differences on pregnancy rates per cycle. They came to the conclusion that HVC had no effect upon IVF treatment or pregnancy.
--Mark Hill (talk) 10:36, 4 September 2015 (AEST)These are good summaries of the 2 papers. You did not need to keep "look at this " at the front of the reference code. (5/5)
Lab 2 Assessment
Upload a research image using the guide information below. The image uploaded for your individual assessment can relate to your project or from fertilisation to week 3 of development (upload only a single image).
Add that image to your own individual page (see Images) including an image title and its reference link.
| Uploading Images in 5 Easy Steps | ||
|---|---|---|
First Read the help page Images and Copyright Tutorial.
Students cannot delete images once uploaded. You will need to email me with the full image name and request deletion, that I am happy to do with no penalty if done before I assess. Non-Table version of this page
|
look at this [3]
PMID 26250560
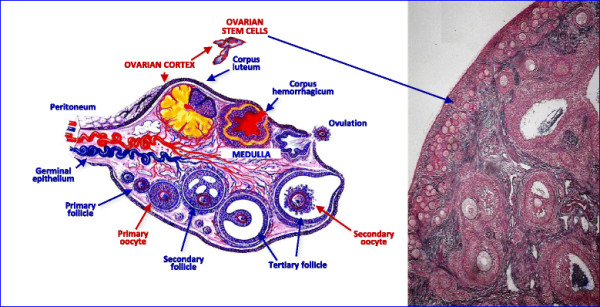
Ovarian stem Cell Schematic [4]
PMID 26250560
--Mark Hill (talk) 10:42, 4 September 2015 (AEST) Image uploaded correctly with reference, copyright and student template. The uploaded image size is so small the text cannot be read, you should have used a larger version of the original figure. Just a few minor issues. The Image title (Ovarian Structural scematic and stained cortical region of an ovariana stem cell .jpg) is far too long and includes an unnecessary space at the end. This could have been (Ovarian stem cell cartoon.jpg). Your subheadings have not been updated/formatted correctly and you have not formatted the reference correctly (I have fixed the reference this please see the difference). While all these are minor, it shows you are not reviewing your work.
(4/5)
Lab 3 Assessment
In Lab 3 you were asked to divide the group project among your members and to research your allocated project component.
You will need to demonstrate that you have actively researched your project topic over the last week by having your section heading appear on the project page and have at least 3 research/review articles listed under that sub-heading.
The same content should be passed on your own individual assessment page.
Prenatal Diagnosis
Prenatal diagnosis is the screening process that tests an early fetus for overall growth, complications of pregnancy, birth defects and chromosomal or genetic abnormalities within the first 2 trimesters. It aims to provide the parents with as information as possible to help them make an informed decision about the infants quality of life. In 90-95% of the cases negative outcomes occur; confirming the healthy state of the fetus, should a genetic abnormality be present, it provides the parents with the opportunity to investigate further with other tests, and possible fetal therapeutic treatments available as well plan and prepare for the disabled infant or to terminate the pregnancy.
Post Implantation Genetic Diagnosis
- Indicators for prenatal screening/ high risk factors include:
- maternal ages > 35 years
- paternal ages > 50-55
- history of 2+ miscarriages
- previous pregnancy or family history of a preexisting genetic or chromosomal disorder
- suspected carriers of genetic disorders
- maternal disease/condition present (high BP, diabetes)
- abnormal ultrasound or serum test results within the first 2 trimesters
- family history of neural tube or other birth defects
- Ultrasonography:
- high frequency sound waves are used to generate a image of the fetus
- relatively non-invasion; its conducted transabdominally or transvaginally ( producing a higher resolution image)
- It reveals the presence/absence of congenital abnormalities, characteristics of fetal growth and development, uterine development status; amount of **amniotic fluid, placental position, umbilical blood flow and the presence of multiple gestation.
- (if abnormalities are detected further testing is recommended)
- Amniotic Fluid Analysis/:
- samples are obtained through amniocentesis
- the amniotic fluid is analyzed for its biochemical composition
- earlier in the pregnancy it can be examined for sex determination and to diagnose genetic or chromosomal disorders present.
- Later into the pregnancy it provides an indication of fetal maturity and well being
- Feta; cells recovered from the amniotic fluid can be cultured for specific karyotypes, to test for Chromosomal Abnormalities, and analysed for Alpha- fetoprotein (AFP) a biochemical marker of metabolic disorders and neural tube defects as well as other abnormalities.
- Amniocentesis- occurs usually between weeks 14-20 as amniotic fluid has reached the optimal volume (150-250mls) allowing 20-30mls to be removed with a relatively low risk or fetal or maternal complication, and in time for a 2nd trimester abortion.
- early amniocentesis ( before week 13) increase the risk of fetal loss, leakage of essential amniotic fluid and talipes equinovarus.
- Chronic Villus Sampling (CVS):
- occurs roughly 10-13 weeks after last menstrual cycle, ensuing a sufficiently developed chorionic villi but before the chorion laeve forms the definitive placenta.
- ultrasounds is used to locate the gestational sac and implantation then a transcervial or transabdominal approach is used to aspirate living tropoblast tissue.
- sample is analyzed for chromosomal abnormalities or with enzyme assay.
- advantages: earlier diagnosis , decreased waiting period
- disadvantages: risk of spontaneous abortion, bacterial infection, bleeding, leakage of amniotic fluid, inability to diagnose neural tube defects this early, early cleavage (before wk 10) is associated with increased risk of limb defects (due to insufficiently developed chronic villi)
- Umbilical Blood Sampling:
- can occur as early as 16 weeks
- using the umbilical cord to obtain fetal blood samples - with real time ultrasound
- used to diagnose inherited blood disorders, to detect congenital infections, to assess fetal anemia and in treatments such as blood transfusions.
- disadvantages: risks of infection, preterm labor, thrombosis, bleeding & transient fetal arrhythmia
- Fluroscent in Situ Hybridisation (FISH)
- rapidly detects (within 24 hours of testing ) the presence of Trisomies 21, 13 and 8 and alterations in sex chromosomes in uncultured cells.
fetal therapies
early diagnosis allows the opportunity for intervention:
- surgical intervention urinary tract obstruction ; aiming to reduce prenatal renal damage
- fetal transfusions ( feta anemia
- fetal medical treatmetn ( fetal cardiac arrhythmias,impaired thyroid function etc.) treatment occurs usually through the mother
- infusions for hematologic conditions
- stem cell transplantation
- gene therapy
- pharmocolic interventions
textbooks used :
- Maternal, Fetal & Neonatal physiology: a Clinical perspective [5]
- Langman's Medical Embryology (12th ed.)Chapter 9, pages 125-129 [6]
the following are two articles about a newly available and accessible prenatal non- invasive genetic test- Blood sampling:
- Report on Cutting edge prenatal screening technology to become available in Australia [7]
- Blood Test takes risk out of prenatal testing [8]
- Prenatal screening and Diagnostic Tests Information Pamphlet [9]
<pubmed>26168107</pubmed>
--Mark Hill (talk) 10:46, 4 September 2015 (AEST) While you have included a lot of content for your group project, I only see a single PubMed reference here? The other sources are useful, but begin with the research literature. (4/5)
Lab 4 Assessment
ANAT2341 Quiz Example | Category:Quiz | ANAT2341 Student 2015 Quiz Questions |
Design 3 quiz questions based upon any of the major subjects we have covered to date. Add the quiz to your own page under Lab 4 assessment and provide a sub-sub-heading on the topic of the quiz. An example is shown below (open this page in view code or edit mode).
Gamatogenesis
Fertilisation and gamatogenesis
ANAT2341 Lab 1 - Gametogenesis
- neither gamate has reached its full maturity when it leaves the gonad - spermatogenesis can not occur untill puberty and oogenesis is reactivated at puberty due to hormual stimulation from the endocrine system
Here are some further questions on fertilisation and Gamatogenesis Quiz 1
--Mark Hill (talk) 10:51, 4 September 2015 (AEST) These are indeed some thought provoking questions you have devised here. You could also have included links to pages where the student could read more about the topics. (9/10)
ANAT2341 Student 2015 Quiz Questions
Lab 5 Assessment
Select one of the topics shown below and write 3 paragraphs (with referenced sources) on that specific topic.
Cleft Lip and cleft palate are associated with many different environmental and genetic causes. Identify and describe one cause of these abnormalities.
Discuss how aganglionic colon is a gastrointestinal tract abnormality related to neural crest migration.
What is the difference between gastroschisis and omphalocele?
During regular embryonic development, the mid gut herniates the abdominal wall (week 6) and develops externally in a sac, ventral to the body wall (week 6-10). Once the fetus has grown to such a caliber that it is large enough and strong enough it recaptures the mid gut through the umbilical ring (week 11), it then rotates internally to reach its correct adult anatomical position. [10] The midgut herniation is a possible site for abnormalities like Gastroschisis or Omphalocele to occur, due to congenital defects of the abdominal wall [11] or abnormal rotation and fixation of the midgut.
Gastroschisis which is the abnormality where the herniated abdominal viscera (midgut) sit to the right of the intact umbilical cord on the outer anterior surface of the body wall, with no membranous covering, the viscera is susceptible to organ damage as it is directly exposed with the amniotic fluid. [12] Although these are not abnormalities of the gastrointestinal system, it has a direct effect upon the development and functions of the GIT causing inflammation of the bowel and delayed alimentation. There are 2.5 cases/10,000 births, which is increasing incidence [13]. Gastroschisis usually occurs in isolation but can be associated associated with young maternal age (< 20 years), folic acid deficiency as well as low gravida, prematurity, and low birth-weight babies, [14] secondary to in utero growth retardation; fetal, Gastrointestinal anomalies (intestinal atresia, perforation, necrosis or volvulus), Hirschsprung's disease, Intestinal Atresia, GERD, Cryptorchidism and sporadically Oromandibular limb hypogenesis. Complications include resulting defects in other organs in 35% of cases and fetal distress. [15] There are several possible theories explaining the cause of this abnormality; when the yolk sac and asscoiated vitelline structures are not included in the umbilical stalk, [16], due to a compromise in the vasculature of the right abdominal wall. [17] Gastroschisis can be simply detected using ultrasound scan and has a relatively high mortality rate of 40%.
The Omphalocele, like Gastroschisis is an abnormailty that is caused by a congenital defect of the abdonminal wall,when the mid herniates the abdominal cavity, unlike the herniated Gastroschisis, the viscera is contained within the umbilical cord (unless it ruptures) and the organs are protected from the amniotic cavity. [18] It is caused by a failure to recapture the herniated midgut in week 11. As long as the herniation is enclosed within a membrane (unless perforated) the midgut should develop and function normally. This condition does occur in association with increased maternal age, twins, consecutive children and different generations of the same family and as a result of with as a result of chromosomal disorders (25-50 % of cases) trisomy 13,14,14,18,21 and also with Beckwith-Wiedemann Syndrome, Macrosomia, GERD Cryptorchidism Musculoskeletal Neural tube defects and congenital heart defects.[19]. Omphalocele and Ganstroschisis are similar conditions with a few essential points of difference.
summary:
Gastroschisis:
Not covered by membrane directly impacts GIT function impaired GIT function herniation lies to the right of umbilical cord occurs isolation and less often with associated conditions higher risk of complication Lower mortality rate, 60% survival rate Larger rate of incidence Unsure of origin- there are several theories easy prenatal diagnosis variable postnatal outcomes.
omphalocele'
Covered by membrane indirectly impacts GIT normal GIT function Herniation within umbilical cord linked with chrosomal abnormalities and other conditions Trisomy 13,14,14,18,21 Higher risk of complication originates in failure to retract the loops of midgut
Lab 6 assessment
History
Historically prenatal genetic diagnosis were conducted post-implantation in the first 2 trimesters, testing for overall fetal growth, complications of pregnancy, birth defects and chromosomal or genetic abnormalities, although, currently these genetic tests are conducted as a part of PreImplantation Genetic Diagnosis (PGD) made possible by the recent advances in Assisted Reproductive Technologies (ART).
Genetic Techniques
main indicaators of the presence single gene disorders (PCR) and inherited chromosome abnormalities(FISH) PMID 25154779 (might be useful?)
Fluorescent In Situ Hybridisation (FISH)
Fluorescence in situ hybridization (FISH) the one of the most effective and rapid [20] Preimplantation Genetic Diagnosis (PGD) techniques used to locate specific DNA sequences within a chromosome. FISH facilitates the clinical diagnosis of chromosomal abnormalities indicated by sequential duplications, deletions and rearrangements of chromosome, which are usually missed with microscopic analysis. This technique is especially relevant for female embryos with X-linked diseases,[21] that have no other mutation specific tests. (http://www.ncbi.nlm.nih.gov/pubmed/20809319) As part of PGD samples are collected from various stages of the embyro, with the test being conducted on the blastocyst, polar body biopsy and blastomeres.[22] FISH is 99% effective when used in conjunction with competitive Genomic Hybridisation (CGH) in diagnosing chromosomal abnormalities.[23]
How does it work ?
Probes are short single strands of DNA that have been tagged with fluorescent labels to complement specific parts of each chromosome. These probes are usually target specific or used in a specific combination to test for specific loci.
These fluorescent labels are small chemical agents that glow brightly in the presence of a specific region on a chromosome.
Heating the DNA sample denatures it, causing the individual DNA strands to break apart, allowing the probe to hybridise (join with) the complementary strand of DNA. the fluorescent tags enable the researchers to correctly identify the presence or lack thereof and location of the specific chromosomes that they are testing for. [24]
The probe will not hybridise is the there has been a duplication or a deletion of the DNA - indicating chromosomal abnormalities such as trisomies 13,18 and 21
PMID: 26338801 PMID: 11325751 PMID: PMID: 11325751 PMC1120169 PMID: 17970921
Unique, Rare Chromosome Disorder Support Group (2013) Fluorescence in situ hybridisation (FISH) Rarechromo.org retrieved from [23]
NIS (2015) Flourescence in Situ Hydridization (FISH) National Human Genome Research Institute retrieved from [24]
Lab 7 assessment
1. Identify and write a brief description of the findings of a recent research paper on development of one of the endocrine organs covered in today's practical.
Proteomic characterization of adrenal gland embryonic development reveals early initiation of steroid metabolism and reduction of the retinoic acid pathway.[25]
The molecular mechanism of adrenal Gland development are not well understood or documented. The purpose of this study was to identify the protein pathways that initiate the specification of the endocrine function in the adrenal gland and provide a comprehensive proteome analysis for 3 separate stages. Electrophoresis ad mass spectrometry analysis were use to monitor the alterations of proteome on at embryonic days (E) E16 and E19 and post natal day 1 P1. They found that the key proteins of cholesterol synthesis increased expression significantly at E19 showing the initiation of endocrine specialization and, that retinoic acid pathways decreased during adrenal gland development , possibly indicating its role in the early stage development.
(More ? Adrenal Development )
2. Identify the embryonic layers and tissues that contribute to the developing teeth.
Odontogenesis or tooth development initiates in week 6 of embryonic development and is contributed to the following cells and tissue layers:
Odontoblasts: are ectomesenchyme cells derived from the neurocrest,. They initially form predentin that calcifies to form dentin, this dentogenesis is driven by the influence of enamel epithelium. (week14)
Ameloblasts: are a derivative of epithelium from the ectoderm cells of the first pharyngeal arch. They produce enamel which covers the crown of the tooth, and are only present during odotogenesis. (week 14)
Peridontal Ligament is a specialized connective tissue containing Shapey's Fibres (specialized collagen fibre bundles) that holds the tooth in place in its socket. It functions as a shock absorber transmitting the chewing forces , as well as relays sensory information like heat, cold, pain and pressure.
Odontogenesis has 4 key stages: bud (week 8), cap (week 11), bell (Week 14) and terminal differentiation.
(Tooth Development UNSW embryology)
<pubmed> 18671204 </pubmed>
L8 assessment
History
Prenatal Genetic Diagnosis was historically conducted post-implantation in the first 2 trimesters [26], testing for overall fetal growth, complications of pregnancy, birth defects as well as chromosomal or genetic abnormalities[27]. Due to advancements in Artificial Reproductive Technologies ( ART) in the past 2 decades , these genetic tests are now conducted as a part of Preimplantation Genetic Diagnosis (PGD) [28] and art used to eliminate the rsik of passing on a genetic disorder to an offspring, before implantation, it ha now become a preventative measure. . In 1990 Preimplantation genetic testing was first used in 1990 on humans to diagnose recessive X-linked diseases in the embryos of pregnant women undergoing IVF. [29] PGD is offered as an alternative to prenatal diagnosis in detecting genetic disorders in high risk couples [30] and avoid passing an inherited disease to the offspring, by selecting genetically "normal" embryos for implantation ,to give the best chance of successful implantation , stable pregnancies and healthy children. [31]
Genetic Techniques
PCR
Description
Polymerase chain reaction (PCR) amplifies DNA specific to genetic sequence of interest . PCR was developed by Kay Mullis in the 1980s, for which he was awarded the Nobel prize for chemistry in 1993. [32] This technique enables clinicians to monitor and diagnose diseases using minute samples such as embryonic cells, blood & tissue. [33] PCR has many important applications like DNA fingerprinting, genetic mapping, detection of viruses and bacteria as well as being used to detect genetic disorders, as a part of PGD, in conjunction with IVF. It is used to detect molecular abnormalities such as single gene disorders like Tay Sach, Cycstic fibrosis, Duchenne Muscular Dystrophy, Thalassemia, Huntington disease, Spinal muscular atrophy and many more. Molecular and Genetic analysis require a significant amount of DNA, which we would not have without PCR. It revolutionized the study of DNA, replacing all previous recombinant DNA technology. [34]
Procedure
Sample are obtained from the the blastocyst, a polar body biopsy or the blastomeres stages of the embryo. Stage 1 Denaturing: separating the target strands of DNA The obtained sample is heated to roughly 90 degrees celcius, this heat causes breaks the relatively weak bonds between nucleotides that form DNA. the double stranded DNA to split into 2 single strands of DNA that are used as templates. Stage 2 Annealing: Binding the Primers to the target DNA sequence PCR will only copy the target sequence of DNA specified by specific PCR primer. These synthesized primers Oligonucleotides are small artificial pieces of DNA. TAQ polymerase enzyme synthesize 2 new strands of DNA duplicate to the single sample DNA stand template indicated by these primers. During this stage the reaction is cooled to a temperature between 40-60 degrees Celsius. Stage 3- Extension- making copies Each of these 2 copies are then used again as templates generating 2 further replications This cycle can occur as many 30 -40 times within a couple hours leading to billions of extra copies of the original DNA segment. The optimal temperature for the further replication is roughly 72 degrees Celsius.
| PCR Cycle | Target Copies |
| 1 | 2 |
| 2 | 4 |
| 3 | 8 |
| 4 | 16 |
| 5 | 32 |
| 6 | 64 |
| 7 | 128 |
| 8 | 256 |
| 9 | 512 |
| 10 | 1024 |
| 15 | 32,768 |
| 20 | 1,048,578 |
| 25 | 33,554,432 |
| 30 | 1,073,741,842 |
This process mediated by a thermocycler machine that is programmed to alter the temperature of the reaction every couple of minutes, perpetuating the cycle of DNA denaturing and synthesis.
This process, generates exponential exact copy of the original template DNA sequence.
Advantages & Disadvantages
ADVANTAGES
- fast and inexpensive way of copying a target sequence of DNA
- aids the diagnosis of single cell defects
- rapid generation of results within a couple hours
- can start from a single molecule of DNA, therefore is more sensitive
- has a flexible primer design enabling
- overcame previous issues associated with the limited availability of sites of end
- generates the DNA copies exponentially
- enables DNA amplification required for molecular and genetic analysis
- Valid diagnostic method in selecting unaffected embryos for embryo transfer [35] [36]
DISADVANTAGES
- only during this exponential DNA replication phase can we determine the starting quantity sequence contained in the original sample ( template DNA strand)
- PCR reaction is limited by the presence of inhibitors present in the sample :
reagent inhibitors
- self annealing due to the accumulation of the product -stopping exponential amplification for the target sequence and reach a plateau
quantification of the end point of reaction of PCR products unreliable - therefore that is why we require real time quantitative RT-PCR necessary [37]
Fluorescent In Situ Hybridisation (FISH)
Description
Fluorescence in situ hybridization (FISH) the one of the most effective and rapid [38] Preimplantation Genetic Diagnosis (PGD). This techniques used to locate a specific DNA sequences within a chromosome. FISH facilitates the clinical diagnosis of chromosomal abnormalities indicated by sequential duplications, deletions and rearrangements of chromosome, that are usually missed with microscopic analysis. This technique is especially relevant for female embryos with X-linked diseases,[39] that have no other mutation specific tests [25] As part of PGD, samples are collected from various stages of the embyro,and they are able to conducted tests on the blastocyst, a polar body biopsy and the blastomeres [40]. FISH is 99% effective when used in conjunction with competitive Genomic Hybridisation (CGH) to diagnose chromosomal abnormalities.[41]
Procedure
Samples are collected from either the blastocyst, a polar body biopsy or the blastomeres stages of the embryo [42]. DNA strands are heated and denatured causing their the individual DNA strands to break apart Probes are single complimentary stands of DNA a that have been tagged with small chemical agents that glow brightly in the presence of a specific region on a chromosome. These specific probes then hybridize and join to their complementary DNA strand. The fluorescent tags enable the researchers to correctly identify the presence or lack thereof and location of the specific chromosomes that they are testing for. [43] Researcher analyse the results by identifying the number and the relative location of the fluorescent dots generated by the FISH images.[44] The probe will not fully hybridise if there has been a duplication or a deletion of the DNA - indicating chromosomal and sex chromosomal anomalies like trisomies and aneuploidies. [45]
Different probes are used for different purposes: [46]
- Locus specific tags detect very small imbalances, and locate isolated small portions
of genes within a chromosome
- Alphoid/Centomeric Repeat probes are developed from the repetitive sequence located in the middle region of each chromosome, useful in determining the number of chromosomes , detecting rearrangement and when used in conjunction with locus probes to determine the absence of genetic material on a chromosome.
- Paint Probes are collections of smaller probes with their own stains that bind to a different section on the chromosome - allowing the full chromosome too be labeled a unique colour, this "full colour map" can be used to know the spectral karyotype- full chromosomal mapping is useful in examining chromosomal abnormalities.
Advantages & Disadvantages
ADVANTAGES
- FISH is 99% effective when used in conjunction with Competitive Genomic Hybridisation (CGH) to diagnose chromosomal abnormalities.[47]
- rapid generation of results
- enables the number and size of specific chromosomes to be known
- identifies very small translocations and aneuploidies that occur within chromosomes, that would usually be missed under microscopic analysis
- indicates whether the accessed chromosomes are normal [48]
- tests for the most common chromosomal abnormalities - down syndrome chromosomes 13,16,18,21 and 22 [49]
- can be used to identify and remove inheritable X-linked disease or sex chromasome anomalies such as Duchennes Muscular Dystrophy, hemophilia, ectodermal dysplasia [50]
- it may be used to compare the chromosomal gene arrangements in related species
DISADVANTAGES
- FISH destroys all cells tested [51]
- it does not fully access all chromosomes ( only ~ 12),
- there is a 40 % chance that chromosomal aneuploidy is occuring and not being targeted by this test [52]
- analysis of results is dependent upon the dot quality impacted by hybridisation efficiency or the camera sensitivity [53]
- some studies indicate that using FISH on a day-3 embryo biopsy decreases the rate of live births [54]
PMID 11325751
Array Comparative Genomic Hybridisation (aCGH)
Description
aCGH also known as Microarray analysis efficiently scans the entire genome for chromosomal imbalances. CGH was initially developed to detect the number of changes in a solid tumor mass. It uses 2 genomes comparing the sample to the control, with each labeled in a different fluorescent dye[55]. Earlier CGH techniques were limited by the resolution of the imaging [56] , these initial limitations were overcome by using Microarrays in conjunction with CGh to improve the resolution of the imaging, Array Comparative Genomic Hybridisation (aCGH). This method compares sample and control microarrayed slides containing small segments of DNA (probes). [57] the Probes used will vary according from the small (25-85 base pairs) oligonucleotides manufactured to highlight different target sequences, to the very large genomic clones (80,000- 200,00 base pairs), and as these are significantly smaller than the traditional metaphase chromosomes used for CGH, generating a higher resolution of image. [58].aCGH is used as a diagnostic tool for prenatal detection f chromosomal abnormalities [59].
Procedure
- sample is obtained (skin, blood or fetal cells) and DNA is obtained.
As a part of PGD fetal cell samples are collected from; the fertilized egg polar bodies, the blastomere (day 3 embryo) or the blastocyst/tropoectoderm stage (day 5 embryo).
- Sample DNA is labeled with one fluorescent dye, and the control DNA is labeled with a different colored fluorescent dye. the control DNA is used as the base point of reference.
heated and denatured single DNA strands then hybridize to their complementary single strand probes
- these are then combined and applied to a microarray
- the results are run through a computer program and a digital imaging system is used to quantify the results( fluorescent intensities of the labeled probes).
simple explanation of aCGH : [Genomics Education (2014) Genetic Testing for Health: aCGH|https://vimeo.com/84757281] [61]
The fluorescent ratio and the hybridization signal at different locations on the genome of the control DNA are used to identify any variances present in the sample DNA,. aCGH facilitates the clinical diagnosis of submicroscopic chromosomal duplication, deletion and rearrangements indicative of chromosomal disorders such as trisomies 1-22and specific sex linked disorders.
Duplications in the DNA are displayed by the computer program as spikes/ peaks over an established threshold and deletions in DNA are displayed by the computer program as spikes/ toughs beneath this threshold
-computer screening image-
-drawn diagram of method-
Advantages & Disadvantages
ADVANTAGES:
- multiple applications: prenatal genetic diagnosis, cancer diagnosis, [62] genetic screening for developmental delay (learning disabilities) [63]. or congenital anomalies that are suspected to be genetic in origin [64]
- trace represents all chromosomes present in the human genome chromosomes 1-22 and the X & Y chromosomes, and therefore is the most accurate method for testing whole embryo aneuploidy
- aCGH has been extensively tested, and Validated, and is now used world wide.
- detects deletions and additions and rearrangements as well as amplification, of the WHOLE genome, simultaneously.
- detects submicroscopic alterations
- used as a diagnostic tool for prenatal detection of chromosomal abnormalities [65].
- provides high resolution genomewide screening of segmental genomic copy number variations
- very accurate when used in conjunction with FISH
- Alone is more reliable and in detecting chromosomal abnormalities, with a higher implantation success rate, than FISH [66].
- Allows an in depth research focus upon specific types of rearrangements within selected chromosomal regions, a recent particular area of interest is subtelomeric and pericentromeric rearrangements
- reduces the risk of failed implantation and miscarriage, improving the chance of a healthy baby [67]
DISADVANTAGES:
- translocations and inversions of DNA are not detected
- limited ability diagnosing specific polyploidies such as triploidy [68]
- will not detect mosaicism detection <20% (cultures where >1 of 5 cells are trisomy 12)
- will not detect balanced chromosomal rearrangement
- will not detect duplications or deletions <80kb
- will not detect point mutations within genes [69]
- will not detect chromosomal position of genomic gains
- will not detect loos of heterozygosity (LOH) or Absence of heterozygosity (AOH) [70]
Next Generation Sequencing
Description
The development and advances within ART in the past 20 years, as well as the increasing popularity of IV, has lead to an influx of new technologies developed to screen embryos for chromosomal anomalies, which are covered by the umbrella term of Next generation Sequencing (NGS). NGS is a general term used to describe all of the new and emerging screening techniques currently being introduced and used as part of PGD for IVF . NGS screens for single gene disorders as well as conducting extensive and very comprehensive chromosome diagnosis by sequencing, counting, and accurately assembling millions of DNA reads, simultaneously. [71] There is a movementfor NGS to replace the other limited testing techniques and be used as the standard.
Procedure
Samples are collected from either blastomere or the blastocyst/tropoectoderm and processed for analysis by a computer system. The methodology of each process is unique to the technique being used.
Advantages & Disadvantages
ADVANTAGES:
- cheaper
- opens new diagnostic possibilities
- accurately tests all 24 chromosomes
- tests for the presence of monogenic diseases of known genetic background
- reduces the number of biopsies required for diagnosis
- higher detection rate of small translocations
- highly accurate is testing for compound point mutations, chromosomal duplication, deletions and insertions [72]
- it accurately detects chromosomal aneuploidy and unbalanced rearrangement [73]
- NGS single gene disorder screenings can conducted in conjunction with PCR comprehensive chromosomal screening.
- Reduces human error
- better detects the presence of mosaicism
- work well in conjunction with CGH and aCGH as part of PGD, improving the chances of IVF.
DISADVANTAGES: there is limited information available to clinical applications of NGS [75]
Lab Attendance
--Z5020317 (talk) 13:46, 7 August 2015 (AEST)
--Z5020317 (talk) 13:15, 14 August 2015 (AEST)
--Z5020317 (talk) 13:51, 21 August 2015 (AEST)- present but forgot to sign in
--Z5020317 (talk) 12:43, 28 August 2015 (AEST)- present but forgot to sign in
--Z5020317 (talk) 13:20, 4 September 2015 (AEST)
--Z5020317 (talk) 12:54, 11 September 2015 (AEST)
--Z5020317 (talk) 12:43, 25 September 2015 (AEST) --Z5020317 (talk) 11:45, 9 October 2015 (AEDT)
References
- ↑ <pubmed>26166637</pubmed>
- ↑ <pubmed>26131230</pubmed>
- ↑ <pubmed>26250560</pubmed>
- ↑ <pubmed>26250560</pubmed>| [1]
- ↑ Blackburn, S.L. (2003) Maternal, Fetal & Neonatal physiology: a Clinical perspective (2nd ed.). Seattle: Saudners
- ↑ Sadler T.W.(2012) Langman's Medical Embryology (12th ed.) Philadelphia: Lipincott, Wiliams & Wilkins, a Wolters Kluwer Business
- ↑ Carbonell, R. Shinners, A. Amor, D. Mark, D. (2015) Report on Cutting edge prenatal screening technology to become available in Australia: Prepared for ABC news, PM with Mark Colvin. Retrieved from {http://www.abc.net.au/pm/content/2015/s4203200.htm}
- ↑ Begley, S. (05/07/2015) Blood Test takes risk out of prenatal testing. ABC Science. Retrieved from {http://www.abc.net.au/science/articles/2012/07/05/3539549.htm}
- ↑ Western Australia. Department of Health Genetics Council Prenatal Diagnosis Committee (2011)Prenatal screening and Diagnostic Tests. Retrieved from {http://www.health.wa.gov.au/docreg/Education/Prevention/Genetics/HP3131_prenatal.pdf}
- ↑ Hill, M.A. (2015) Embryology Lecture - Gastrointestinal Development. Retrieved September 11, 2015, from https://embryology.med.unsw.edu.au/embryology/index.php/Lecture_-_Gastrointestinal_Development
- ↑ <pubmed>12557047</pubmed>
- ↑ Hill, M.A. (2015) Embryology Musculoskeletal System - Abnormalities. Retrieved September 11, 2015, from [2]
- ↑ <pubmed> 16563666</pubmed>
- ↑ <pubmed> 22004141 </pubmed>| BMC Pediatr.
- ↑ <pubmed>15488122</pubmed>
- ↑ <pubmed>19419415</pubmed>
- ↑ Boykin, Kevin Dr Gastroschisis vs Omphalocele; Pediatric SurgeryLSU Health and Science Center Shreveport PDF retrieved from [3]
- ↑ Hill, M.A. (2015) Embryology Gastrointestinal Tract - Abnormalities. Retrieved September 11, 2015, from https://embryology.med.unsw.edu.au/embryology/index.php/Gastrointestinal_Tract_-_Abnormalities
- ↑ Glasser J. G., MD, MA, FACS , Rosenkrantz T., MD (2015, April 28) Pediatric Omphalocele and Gastroschisis: Background, Pathophysiology, Epidemiology Medscape retrieved from [4]
- ↑ <pubmed>26338801</pubmed>
- ↑ <pubmed>20809319</pubmed>
- ↑ <pubmed>21748341</pubmed>
- ↑ <pubmed>26338801</pubmed>
- ↑ <pubmed>17876073</pubmed>
- ↑ <pubmed> 25694770</pubmed>
- ↑ Blackburn, S.L. (2003) Maternal, Fetal & Neonatal physiology: a Clinical perspective (2nd ed.). Seattle: Saudners
- ↑ Sadler T.W.(2012) Langman's Medical Embryology (12th ed.) Philadelphia: Lipincott, Wiliams & Wilkins, a Wolters Kluwer Business
- ↑ <pubmed>20638568</pubmed>
- ↑ <pubmed>2330030</pubmed>
- ↑ <pubmed>26168107</pubmed>
- ↑ <pubmed>23499002</pubmed>
- ↑ Pubmed Docs (2015) Polymerase Chain Reaction (PCR) Pubmen. Retrieved from [5]
- ↑ Roche (2015) PCR: How We Copy DNA. Roche Molecular Systems Inc. retrieved From [6]
- ↑ Mullins, K., Francois, F.& Gibbs R.A. (1994 ) The Polymerase Chain Reaction. p3. Springer- Science +Business Media. Birkhauser, Boston Available at [7]
- ↑ Dreesen, J., Drusedaul, M., Smeets, H., Die-Smulders, C., Coonen, E., Dumoulin, J., Gielen, M., Evers, J., Herbergers, J. & Geradets, J. (2008) Validation of preimplantation genetic diagnosis by PCR analysis: genotype comparison of the blastomere and corresponding embryo, implications for clinical practice. Mol. Hum. Reprod. 14 (10):573-579.doi: 10.1093/molehr/gan052. Retrieved from [8]
- ↑ <pubmed>20966460</pubmed>
- ↑ NIS (2015) Polymerase Chain Reaction (PCR) National Human Genome Research Institute retrieved from [9]
- ↑ <pubmed>26338801</pubmed>
- ↑ <pubmed>20809319</pubmed>
- ↑ <pubmed>21748341</pubmed>
- ↑ <pubmed>26338801</pubmed>
- ↑ <pubmed>21748341</pubmed>
- ↑ <pubmed>17876073</pubmed>
- ↑ <pubmed>17970921</pubmed>
- ↑ NIS (2015) Flourescent in Situ Hydridization (FISH) National Human Genome Research Institute retrieved from [10]
- ↑ Unique, Rare Chromosome Disorder Support Group (2013) Fluorescence in situ hybridisation (FISH) Rarechromo.org retrieved from [11]
- ↑ <pubmed>26338801</pubmed>
- ↑ Iyer, B. (2013) SlideShare PreImplantation genetic diagnosis(pgd) Retrieved Oct 2, 2015 [12]
- ↑ <pubmed>21749752</pubmed>
- ↑ <pubmed>17876073</pubmed>
- ↑ O'Connor, C. (2008) Fluorescence in situ hybridization (FISH). Nature Education 1(1):171. retrieved from [13]
- ↑ Iyer, B. (2013) SlideShare PreImplantation genetic diagnosis(pgd) Retrieved Oct 2, 2015 [14]
- ↑ <pubmed>17970921</pubmed>
- ↑ <pubmed>26168107</pubmed>
- ↑ <pubmed>1876176</pubmed>
- ↑ Lichter, P., et al. Comparative genomic hybridization: Uses and limitations. Seminars in Hematology 37, 348–357 (2000)
- ↑ Lucito, R., et al. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Research 13, 2291–2305 (2003)
- ↑ <pubmed>22467166</pubmed>
- ↑ <pubmed>19012303</pubmed>
- ↑ Theisen, A. (2008) Microarray-based Comparative Genomic Hybridization (aCGH). Nature Education 1(1):45. Retrieved from [15]
- ↑ Genomics Education (2014) Genetic Testing for Health: aCGH [video file] Vimeo [16]
- ↑ <pubmed>20193845</pubmed>
- ↑ <pubmed>17309648</pubmed>
- ↑ NHS Array Comparitive Genomic Hybridisation (arrayCGH) pgh foundation . retrieved from [17]
- ↑ <pubmed>22034057</pubmed>
- ↑ <pubmed>20494259</pubmed>
- ↑ Iyer, B. (2013) SlideShare PreImplantation genetic diagnosis(pgd) Retrieved Oct 2, 2015 [18]
- ↑ Unique, Rare Chromosome Disorder Support Group (2013) Microarray-based Comparative Genomic Hybridiation (array CGH) Rarechromo.org retrieved from [19]
- ↑ Washington department of education (CGH- FAQ for physicians. Signature Genomic LAboratories, LLC. Retrieved from [20]
- ↑ WiCell Research Institute Inc. (2012) Comparative Genomic Hybridization Microarray. retrieved from [21]
- ↑ <pubmed>23499002</pubmed>
- ↑ <pubmed>23312231</pubmed>
- ↑ <pubmed>25685330</pubmed>
- ↑ Morris, R. S. (2015 ) Next generation sequencing for PCG|PGS|CCS. IVF1 retrieved from [22]
- ↑ <pubmed>26100406</pubmed>
Please do not use your real name on this website, use only your student number.
- 2015 Course: Week 2 Lecture 1 Lecture 2 Lab 1 | Week 3 Lecture 3 Lecture 4 Lab 2 | Week 4 Lecture 5 Lecture 6 Lab 3 | Week 5 Lecture 7 Lecture 8 Lab 4 | Week 6 Lecture 9 Lecture 10 Lab 5 | Week 7 Lecture 11 Lecture 12 Lab 6 | Week 8 Lecture 13 Lecture 14 Lab 7 | Week 9 Lecture 15 Lecture 16 Lab 8 | Week 10 Lecture 17 Lecture 18 Lab 9 | Week 11 Lecture 19 Lecture 20 Lab 10 | Week 12 Lecture 21 Lecture 22 Lab 11 | Week 13 Lecture 23 Lecture 24 Lab 12 | 2015 Projects: Three Person Embryos | Ovarian Hyper-stimulation Syndrome | Polycystic Ovarian Syndrome | Male Infertility | Oncofertility | Preimplantation Genetic Diagnosis | Students | Student Designed Quiz Questions | Moodle page