Non-Invasive Prenatal Testing
| Embryology - 27 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
| Educational Use Only - Embryology is an educational resource for learning concepts in embryological development, no clinical information is provided and content should not be used for any other purpose. |
Introduction
Non-Invasive Prenatal Testing (NIPT) include new techniques that analyzes cell-free fetal DNA circulating in maternal blood or from fetal cells in the cervical canal.
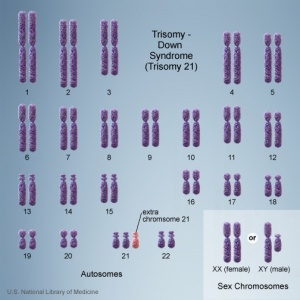
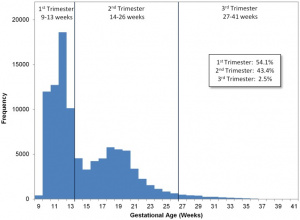
Testing of circulating cell-free fetal DNA (ccffDNA) can be carried out after 10 weeks (between 10-22 weeks) analysis can take a week or more. It has been most useful for replacing amniocentesis in testing for the trisomies; Trisomy 21, Trisomy 18, and Trisomy 13.
There are other pages that refer to postnatal diagnostic testing. (More? Neonatal Diagnosis)
- This Embryology site is a developmental educational resource, it does not provide specific clinical details, you should always refer to a health professional.
| Assisted Reproductive Technology | In Vitro Fertilization | Journal - Prenatal diagnosis
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Non-Invasive Prenatal Testing | Fetal fraction-based risk |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Costs
A recent 2014 Canadian studyPubmedParser error: The PubmedParser extension received invalid XML data. () identified the cost of NIPT ranges from US$800 to US$2000 in the USA and from US$500 to US$1500 elsewhere.
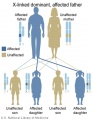
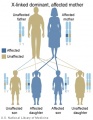
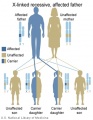
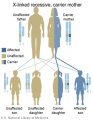
Inheritance Genetics
- Links: Genetic risk maternal age | Trisomy 21
Australia
A recent publication from NHMRC Medical Genetic Testing: information for health professionals (2010). This paper covers background information on all types of genetic tests, not just those associated with prenatal diagnosis.
Types of genetic tests
- Somatic cell genetic testing involves testing tissue (usually cancer) for non-heritable mutations. This may be for diagnostic purposes, or to assist in selecting treatment for a known cancer.
- Diagnostic testing for heritable mutations involves testing an affected person to identify the underlying mutation(s) responsible for the disease. This typically involves testing one or more genes for a heritable mutation.
- Predictive testing for heritable mutations involves testing an unaffected person for a germline mutation identified in genetic relatives. The risk of disease will vary according to the gene, the mutation and the family history.
- Carrier testing for heritable mutations involves testing for the presence of a mutation that does not place the person at increased risk of developing the disease, but does increase the risk of having an affected child developing the disease.
- Pharmacogenetic testing for a genetic variant that alters the way a drug is metabolised. These variants can involve somatic cells or germline changes. Even if these variants are heritable (that is germline changes), the tests are usually of relevance to genetic relatives only if they are being treated with the same type of medication.
USA
A new site developed by NIH "GeneTests" provides medical genetics information resources available at no cost to all interested persons. It contains educational information, a directory of genetic testing laboratories and links to other databases such as OMIM.
- Links: GeneTests | Medline Plus - Genetic Testing
Ethics of Testing
Major developmental abnormalities detected early enough can be resolved far more easily than those discovered late in a pregnancy.
What are the ethical questions that are raised by prenatal testing? Future individual rights or parents rights? But what about diseases, like Huntington's, where a diagnostic test can be made but there are no current treatments for the postnatal (95% of cases adult onset) disease?
Huntington's disease
Guidelines for the molecular genetics predictive test
- Recommendation 2.1 "the test is available only to individuals who have reached the age of majority."
- Recommendation 7.2 "the couple requesting antenatal testing must be clearly informed that if they intend to complete the pregnancy if the fetus is a carrier of the gene defect, there is no valid reason for performing the test."
Movies
Panorama Test
<html5media width="480" height="360">https://www.youtube.com/embed/0xnaCdkV89M</html5media>
- Links: YouTube
References
- ↑ McCullough RM, Almasri EA, Guan X, Geis JA, Hicks SC, Mazloom AR, Deciu C, Oeth P, Bombard AT, Paxton B, Dharajiya N & Saldivar JS. (2014). Non-invasive prenatal chromosomal aneuploidy testing--clinical experience: 100,000 clinical samples. PLoS ONE , 9, e109173. PMID: 25289665 DOI.
- ↑ Su JY, Wei YN, Chen HF, Tong JR, Chen Y, Deng L, Huang LL & Zhang LY. (2023). Analysis of the results of non-invasive prenatal testing (NIPT) in 545 pregnant women in advanced maternal age. Eur Rev Med Pharmacol Sci , 27, 7101-7106. PMID: 37606120 DOI.
- ↑ Huang L, Bogale B, Tang Y, Lu S, Xie XS & Racowsky C. (2019). Noninvasive preimplantation genetic testing for aneuploidy in spent medium may be more reliable than trophectoderm biopsy. Proc. Natl. Acad. Sci. U.S.A. , 116, 14105-14112. PMID: 31235575 DOI.
- ↑ Xue Y, Zhao G, Li H, Zhang Q, Lu J, Yu B & Wang T. (2019). Non-invasive prenatal testing to detect chromosome aneuploidies in 57,204 pregnancies. Mol Cytogenet , 12, 29. PMID: 31249627 DOI.
- ↑ McKanna T, Ryan A, Krinshpun S, Kareht S, Marchand K, Grabarits C, Ali M, McElheny A, Gardiner K, LeChien K, Hsu M, Saltzman D, Stosic M, Martin K & Benn P. (2018). Fetal fraction-based risk algorithm for non-invasive prenatal testing: screening for trisomy 13, 18, and triploidy in women with low cell-free fetal DNA. Ultrasound Obstet Gynecol , , . PMID: 30014528 DOI.
- ↑ Jain CV, Kadam L, van Dijk M, Kohan-Ghadr HR, Kilburn BA, Hartman C, Mazzorana V, Visser A, Hertz M, Bolnick AD, Fritz R, Armant DR & Drewlo S. (2016). Fetal genome profiling at 5 weeks of gestation after noninvasive isolation of trophoblast cells from the endocervical canal. Sci Transl Med , 8, 363re4. PMID: 27807286 DOI.
- ↑ Chitty LS, Wright D, Hill M, Verhoef TI, Daley R, Lewis C, Mason S, McKay F, Jenkins L, Howarth A, Cameron L, McEwan A, Fisher J, Kroese M & Morris S. (2016). Uptake, outcomes, and costs of implementing non-invasive prenatal testing for Down's syndrome into NHS maternity care: prospective cohort study in eight diverse maternity units. BMJ , 354, i3426. PMID: 27378786
- ↑ Taneja PA, Prosen TL, de Feo E, Kruglyak KM, Halks-Miller M, Curnow KJ & Bhatt S. (2017). Fetal aneuploidy screening with cell-free DNA in late gestation. J. Matern. Fetal. Neonatal. Med. , 30, 338-342. PMID: 27124739 DOI.
- ↑ Johansen P, Richter SR, Balslev-Harder M, Miltoft CB, Tabor A, Duno M & Kjaergaard S. (2016). Open source non-invasive prenatal testing platform and its performance in a public health laboratory. Prenat. Diagn. , 36, 530-6. PMID: 27027563 DOI.
- ↑ Benn P, Curnow KJ, Chapman S, Michalopoulos SN, Hornberger J & Rabinowitz M. (2015). An Economic Analysis of Cell-Free DNA Non-Invasive Prenatal Testing in the US General Pregnancy Population. PLoS ONE , 10, e0132313. PMID: 26158465 DOI.
- ↑ Chitty LS, Finning K, Wade A, Soothill P, Martin B, Oxenford K, Daniels G & Massey E. (2014). Diagnostic accuracy of routine antenatal determination of fetal RHD status across gestation: population based cohort study. BMJ , 349, g5243. PMID: 25190055
- ↑ Norwitz ER & Levy B. (2013). Noninvasive prenatal testing: the future is now. Rev Obstet Gynecol , 6, 48-62. PMID: 24466384
- ↑ Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, Gammill HS, Rubens CE, Santillan DA, Murray JC, Tabor HK, Bamshad MJ, Eichler EE & Shendure J. (2012). Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med , 4, 137ra76. PMID: 22674554 DOI.
- ↑ Guo X, Bayliss P, Damewood M, Varney J, Ma E, Vallecillo B & Dhallan R. (2012). A noninvasive test to determine paternity in pregnancy. N. Engl. J. Med. , 366, 1743-5. PMID: 22551147 DOI.
Reviews
Jayashankar SS, Nasaruddin ML, Hassan MF, Dasrilsyah RA, Shafiee MN, Ismail NAS & Alias E. (2023). Non-Invasive Prenatal Testing (NIPT): Reliability, Challenges, and Future Directions. Diagnostics (Basel) , 13, . PMID: 37568933 DOI.
Gekas J, Langlois S, Ravitsky V, Audibert F, van den Berg DG, Haidar H & Rousseau F. (2016). Non-invasive prenatal testing for fetal chromosome abnormalities: review of clinical and ethical issues. Appl Clin Genet , 9, 15-26. PMID: 26893576 DOI.
Skirton H, Goldsmith L, Jackson L, Lewis C & Chitty LS. (2015). Non-invasive prenatal testing for aneuploidy: a systematic review of Internet advertising to potential users by commercial companies and private health providers. Prenat. Diagn. , 35, 1167-75. PMID: 26266986 DOI.
Allyse M, Minear MA, Berson E, Sridhar S, Rote M, Hung A & Chandrasekharan S. (2015). Non-invasive prenatal testing: a review of international implementation and challenges. Int J Womens Health , 7, 113-26. PMID: 25653560 DOI.
Benn P. (2014). Non-Invasive Prenatal Testing Using Cell Free DNA in Maternal Plasma: Recent Developments and Future Prospects. J Clin Med , 3, 537-65. PMID: 26237390 DOI.
Articles
Van Opstal D & Srebniak MI. (2016). Cytogenetic confirmation of a positive NIPT result: evidence-based choice between chorionic villus sampling and amniocentesis depending on chromosome aberration. Expert Rev. Mol. Diagn. , 16, 513-20. PMID: 26864482 DOI.
Fairbrother G, Burigo J, Sharon T & Song K. (2016). Prenatal screening for fetal aneuploidies with cell-free DNA in the general pregnancy population: a cost-effectiveness analysis. J. Matern. Fetal. Neonatal. Med. , 29, 1160-4. PMID: 26000626 DOI.
Straver R, Oudejans CB, Sistermans EA & Reinders MJ. (2016). Calculating the fetal fraction for noninvasive prenatal testing based on genome-wide nucleosome profiles. Prenat. Diagn. , 36, 614-21. PMID: 26996738 DOI.
Ohnhaeuser T & Schmitz D. (2016). Non-invasive Prenatal Testing (NIPT): Better Meet an Expert!: The Case of a Late Detected Trisomy 13 Reveals Structural Problems in NIPT Counselling and Highlights Substantial Risks for the Reproductive Autonomy. Geburtshilfe Frauenheilkd , 76, 277-279. PMID: 27064737 DOI.
Orhant L, Anselem O, Fradin M, Becker PH, Beugnet C, Deburgrave N, Tafuri G, Letourneur F, Goffinet F, Allach El Khattabi L, Leturcq F, Bienvenu T, Tsatsaris V & Nectoux J. (2016). Droplet digital PCR combined with minisequencing, a new approach to analyze fetal DNA from maternal blood: application to the non-invasive prenatal diagnosis of achondroplasia. Prenat. Diagn. , 36, 397-406. PMID: 26850935 DOI.
Higuchi EC, Sheldon JP, Zikmund-Fisher BJ & Yashar BM. (2016). Non-invasive prenatal screening for trisomy 21: Consumers' perspectives. Am. J. Med. Genet. A , 170A, 375-85. PMID: 26553705 DOI.
Books
Search Pubmed
Search Pubmed: Non-Invasive Prenatal Testing
- ART - Assisted Reproductive Technology a general term to describe all the clinical techniques used to aid fertility.
- blastomere biopsy - An ART preimplantation genetic diagnosis technique carried out at cleavage stage (day 3), excluding poor quality embryos, detects chromosomal abnormalities of both maternal and paternal origin. May not detect cellular mosaicism in the embryo.
- blastocyst biopsy - An ART preimplantation genetic diagnosis technique carried out at blastocyst stage (day 4-5), removes several trophoblast (trophoderm) cells, detects chromosomal abnormalities of both maternal and paternal origin and may detect cellular mosaicism.
- cell-free fetal deoxyribonucleic acid - (cfDNA) refers to fetal DNA circulating and isolated from the plasma portion of maternal blood. Can be performed from GA 10 weeks as a first-tier test or as a second-tier test, with women with increased probability on combined first trimester screening offered cfDNA or diagnostic testing.
- false negative rate - The proportion of pregnancies that will test negative given that the congenital anomaly is present.
- false positive rate - The proportion of pregnancies that will test positive given that the congenital anomaly is absent.
- free β human chorionic gonadotrophin - beta-hCG subunit of hCG used as a diagnostic marker for: early detection of pregnancy, Trisomy 21, spontaneous abortion, ectopic pregnancy, hydatidiform mole or choriocarcinoma.
- multiples of the median - (MoM) A multiple of the median is a measure of how far an individual test result deviates from the median and is used to report the results of medical screening tests, particularly where the results of the individual tests are highly variable.
- negative predictive value - The probability that a congenital anomaly is absent given that the prenatal screening test is negative.
- Non-Invasive Prenatal Testing - (NIPT) could refer to ultrasound or other imaging techniques, but more frequently used to describe analysis of cell-free fetal DNA circulating in maternal blood.
- polar body biopsy - (PB biopsy) An ART preimplantation genetic diagnosis technique that removes either the first or second polar body from the zygote. As these are generated by oocyte meiosis they detects chromosomal abnormalities only on the female genetics.
- positive predictive value - The probability that a congenital anomaly is present given that the prenatal screening test is positive.
- pre-implantation genetic diagnosis - (PGD, pre-implantation genetic screening) a diagnostic procedure for embryos produced through Assisted Reproductive Technology (ART, in vitro fertilisation, IVF) for genetic diseases that would generate developmental abnormalities or serious postnatal diseases.
- prenatal screening sensitivity - (detection rate) The probability of testing positive on a prenatal screening test if the congenital anomaly is present.
- prenatal screening specificity - The probability of testing negative on a prenatal screening test if the congenital anomaly is absent.
- quadruple test (maternal serum testing of a-fetoprotein Template:AFP, free B-hCG or total hCG, unconjugated estriol, and inhibin A) is a fetal chromosomal anomaly test usually carried out later in pregnancy (GA 14 to 20 weeks).
- single nucleotide polymorphisms - (SNPs) the variation in a single DNA nucleotide that occurs at a specific position in the genome.
- triple test - (maternal serum testing of a-fetoprotein Template:AFP, free B-hCG or total hCG, and unconjugated estriol) is a fetal chromosomal anomaly test usually carried out later in pregnancy (GA 14 to 20 weeks).
| Other Terms Lists |
|---|
| Terms Lists: ART | Birth | Bone | Cardiovascular | Cell Division | Endocrine | Gastrointestinal | Genital | Genetic | Head | Hearing | Heart | Immune | Integumentary | Neonatal | Neural | Oocyte | Palate | Placenta | Radiation | Renal | Respiratory | Spermatozoa | Statistics | Tooth | Ultrasound | Vision | Historic | Drugs | Glossary |
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- Australian and New Zealand
- Royal Australian College of General Practitioners Australian Family Physician (2014)
- College of Obstetricians and Gynaecologists Intrapartum Fetal Surveillance Clinical Guidelines - Second Edition (June 2011)
- IVF Australia prenatal-testing | Panorama Test
- Canada - Society of Obstetricians and Gynaecologists of Canada
- USA
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 27) Embryology Non-Invasive Prenatal Testing. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Non-Invasive_Prenatal_Testing
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G