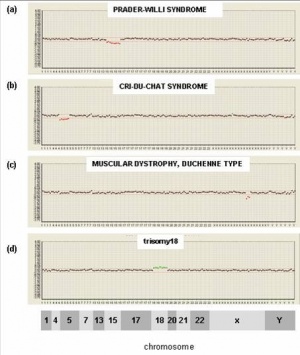
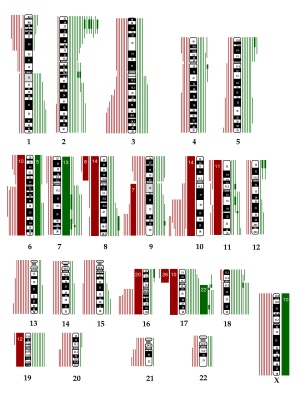
Comparative Genomic Hybridization
| Embryology - 1 May 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
This new test under development is based upon microarray-based comparative genomic hybridization (array CGH). All fetal cells should have complete copies of maternal and paternal genomes. The test compares regions of fetal DNA that deviate from this "pattern" due to either too much or too little DNA, alterations reflect regions of the genome that are either copied or deleted. These genetic changes may therefore cause disease.
Any two unrelated people have the same 99.9% DNA sequences, the remaining 0.1% is the genetic variants that influence how people differ in their risk of disease or their response to drugs.
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Comparative Genomic Hybridization' |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
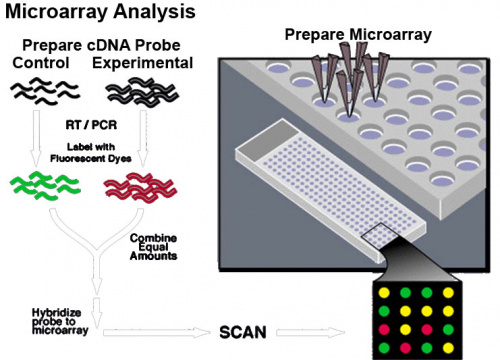
Microarray
Microarrays are a new technique that allows a large number of different genetic sequences to be arrayed on a slide which can then be used to identify specific sequences in an unknown mix of either DNA or RNA. This technique required the development of DNA synthetic techniques and microprocessor controlled robotics for array design and has been used extensively in cell biology.
What is an array? "to place in an orderly arrangement" Think of a hairbrush with each "hair" dipped in individual coloured inks and applied to paper. Each "hair" would makes an individual coloured "spot". Now consider instead of ink, if each hair had a different piece of DNA then a specific pattern is generated.
References
- ↑ Majumdar G, Majumdar A, Lall M, Verma IC & Upadhyaya KC. (2016). Preimplantation genetic screening for all 24 chromosomes by microarray comparative genomic hybridization significantly increases implantation rates and clinical pregnancy rates in patients undergoing in vitro fertilization with poor prognosis. J Hum Reprod Sci , 9, 94-100. PMID: 27382234 DOI.
- ↑ Guo X, Bayliss P, Damewood M, Varney J, Ma E, Vallecillo B & Dhallan R. (2012). A noninvasive test to determine paternity in pregnancy. N. Engl. J. Med. , 366, 1743-5. PMID: 22551147 DOI.
- ↑ Park JH, Woo JH, Shim SH, Yang SJ, Choi YM, Yang KS & Cha DH. (2010). Application of a target array comparative genomic hybridization to prenatal diagnosis. BMC Med. Genet. , 11, 102. PMID: 20576126 DOI.
- ↑ Warren JE, Turok DK, Maxwell TM, Brothman AR & Silver RM. (2009). Array comparative genomic hybridization for genetic evaluation of fetal loss between 10 and 20 weeks of gestation. Obstet Gynecol , 114, 1093-102. PMID: 20168112 DOI.
- ↑ Chinault AC, Shaw CA, Brundage EK, Tang LY & Wong LJ. (2009). Application of dual-genome oligonucleotide array-based comparative genomic hybridization to the molecular diagnosis of mitochondrial DNA deletion and depletion syndromes. Genet. Med. , 11, 518-26. PMID: 19546809 DOI.
- ↑ Nivoloni Kde A, da Silva-Costa SM, Pomílio MC, Pereira T, Lopes Kde C, de Moraes VC, Alexandrino F, de Oliveira CA & Sartorato EL. (2010). Newborn hearing screening and genetic testing in 8974 Brazilian neonates. Int. J. Pediatr. Otorhinolaryngol. , 74, 926-9. PMID: 20538352 DOI.
- ↑ Hardin J, Finnell RH, Wong D, Hogan ME, Horovitz J, Shu J & Shaw GM. (2009). Whole genome microarray analysis, from neonatal blood cards. BMC Genet. , 10, 38. PMID: 19624846 DOI.
- ↑ De Bortoli M, Castellino RC, Lu XY, Deyo J, Sturla LM, Adesina AM, Perlaky L, Pomeroy SL, Lau CC, Man TK, Rao PH & Kim JY. (2006). Medulloblastoma outcome is adversely associated with overexpression of EEF1D, RPL30, and RPS20 on the long arm of chromosome 8. BMC Cancer , 6, 223. PMID: 16968546 DOI.
Reviews
{#pmid:20494259}}
Articles
{#pmid:19795450}}
{#pmid:15127362}}
Search PubMed
Search PubMed: Prenatal Diagnosis Comparative Genomic Hybridization | Comparative Genomic Hybridization
- ART - Assisted Reproductive Technology a general term to describe all the clinical techniques used to aid fertility.
- blastomere biopsy - An ART preimplantation genetic diagnosis technique carried out at cleavage stage (day 3), excluding poor quality embryos, detects chromosomal abnormalities of both maternal and paternal origin. May not detect cellular mosaicism in the embryo.
- blastocyst biopsy - An ART preimplantation genetic diagnosis technique carried out at blastocyst stage (day 4-5), removes several trophoblast (trophoderm) cells, detects chromosomal abnormalities of both maternal and paternal origin and may detect cellular mosaicism.
- cell-free fetal deoxyribonucleic acid - (cfDNA) refers to fetal DNA circulating and isolated from the plasma portion of maternal blood. Can be performed from GA 10 weeks as a first-tier test or as a second-tier test, with women with increased probability on combined first trimester screening offered cfDNA or diagnostic testing.
- false negative rate - The proportion of pregnancies that will test negative given that the congenital anomaly is present.
- false positive rate - The proportion of pregnancies that will test positive given that the congenital anomaly is absent.
- free β human chorionic gonadotrophin - beta-hCG subunit of hCG used as a diagnostic marker for: early detection of pregnancy, Trisomy 21, spontaneous abortion, ectopic pregnancy, hydatidiform mole or choriocarcinoma.
- multiples of the median - (MoM) A multiple of the median is a measure of how far an individual test result deviates from the median and is used to report the results of medical screening tests, particularly where the results of the individual tests are highly variable.
- negative predictive value - The probability that a congenital anomaly is absent given that the prenatal screening test is negative.
- Non-Invasive Prenatal Testing - (NIPT) could refer to ultrasound or other imaging techniques, but more frequently used to describe analysis of cell-free fetal DNA circulating in maternal blood.
- polar body biopsy - (PB biopsy) An ART preimplantation genetic diagnosis technique that removes either the first or second polar body from the zygote. As these are generated by oocyte meiosis they detects chromosomal abnormalities only on the female genetics.
- positive predictive value - The probability that a congenital anomaly is present given that the prenatal screening test is positive.
- pre-implantation genetic diagnosis - (PGD, pre-implantation genetic screening) a diagnostic procedure for embryos produced through Assisted Reproductive Technology (ART, in vitro fertilisation, IVF) for genetic diseases that would generate developmental abnormalities or serious postnatal diseases.
- prenatal screening sensitivity - (detection rate) The probability of testing positive on a prenatal screening test if the congenital anomaly is present.
- prenatal screening specificity - The probability of testing negative on a prenatal screening test if the congenital anomaly is absent.
- quadruple test (maternal serum testing of a-fetoprotein Template:AFP, free B-hCG or total hCG, unconjugated estriol, and inhibin A) is a fetal chromosomal anomaly test usually carried out later in pregnancy (GA 14 to 20 weeks).
- single nucleotide polymorphisms - (SNPs) the variation in a single DNA nucleotide that occurs at a specific position in the genome.
- triple test - (maternal serum testing of a-fetoprotein Template:AFP, free B-hCG or total hCG, and unconjugated estriol) is a fetal chromosomal anomaly test usually carried out later in pregnancy (GA 14 to 20 weeks).
| Other Terms Lists |
|---|
| Terms Lists: ART | Birth | Bone | Cardiovascular | Cell Division | Endocrine | Gastrointestinal | Genital | Genetic | Head | Hearing | Heart | Immune | Integumentary | Neonatal | Neural | Oocyte | Palate | Placenta | Radiation | Renal | Respiratory | Spermatozoa | Statistics | Tooth | Ultrasound | Vision | Historic | Drugs | Glossary |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, May 1) Embryology Comparative Genomic Hybridization. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Comparative_Genomic_Hybridization
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G