Talk:Zebrafish Development
| About Discussion Pages |
|---|
On this website the Discussion Tab or "talk pages" for a topic has been used for several purposes:
Glossary Links
Cite this page: Hill, M.A. (2026, March 1) Embryology Zebrafish Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Talk:Zebrafish_Development |
- This is a timelapse recording of about 18 hours of embryonic development of the zebrafish, Danio rerio, with some annotation added http://www.youtube.com/watch?v=6PhnHYZ5
2023
Novel Development of Magnetic Resonance Imaging to Quantify the Structural Anatomic Growth of Diverse Organs in Adult and Mutant Zebrafish PMID: 37603286
Zebrafish (Danio rerio) is a widely used vertebrate animal for modeling genetic diseases by targeted editing strategies followed by gross phenotypic and biomarker characterization. While larval transparency permits microscopic detection of anatomical defects, histological adult screening for organ-level defects remains invasive, tedious, inefficient, and subject to technical artifact. Here, we describe a noninvasive magnetic resonance imaging (MRI) approach to systematically screen adult zebrafish for anatomical growth defects. An anatomical atlas of wild-type (WT) zebrafish at 5-31 months post-fertilization was created by ex vivo MRI with a 9.4 T magnet. Volumetric growth over time was measured of animals and major organs, including the brain, spinal cord, heart, eyes, optic nerve, ear, liver, kidneys, and swim bladder. Subsequently, surf1-/-, fbxl4-/-, and opa1+/- mitochondrial disease mutant adult zebrafish were quantitatively studied to compare organ volumes with age-matched WT zebrafish. Results demonstrated that MRI enabled noninvasive, high-resolution, rapid screening of mutant adult zebrafish for overall and organ-specific growth abnormalities. Detailed volumetric analyses of three mitochondrial disease mutants delineated specific organ differences, including significantly increased brain growth in surf1-/- and opa1+/-, and marginally significant decreased heart and spinal cord volumes in surf1-/- mutants. This is interesting as we know neurological involvement can be seen in SURF1-/- patients with ataxia, dystonia, and lesions in basal ganglia, as well as in OPA1+/- patients with spasticity, ataxia, and hyperreflexia indicative of neuropathology. Similarly, cardiomyopathy is a known sequelae of cardiac pathology in patients with SURF1-/--related disease. Future studies will define MRI signaling patterns of organ dysfunction to further delineate specific pathology.
2021
ZebraShare: a new venue for rapid dissemination of zebrafish mutant data
PMID: 33954026 PMCID: PMC8051354 DOI: 10.7717/peerj.11007 Free PMC article Abstract
Background: In the past decade, the zebrafish community has widely embraced targeted mutagenesis technologies, resulting in an abundance of mutant lines. While many lines have proven to be useful for investigating gene function, many have also shown no apparent phenotype, or phenotypes not of interest to the originating lab. In order for labs to document and share information about these lines, we have created ZebraShare as a new resource offered within ZFIN.
Methods: ZebraShare involves a form-based submission process generated by ZFIN. The ZebraShare interface (https://zfin.org/action/zebrashare) can be accessed on ZFIN under "Submit Data". Users download the Submission Workbook and complete the required fields, then submit the completed workbook with associated images and captions, generating a new ZFIN publication record. ZFIN curators add the submitted phenotype and mutant information to the ZFIN database, provide mapping information about mutations, and cross reference this information across the appropriate ZFIN databases. We present here examples of ZebraShare submissions, including phf21aa, kdm1a, ctnnd1, snu13a, and snu13b mutant lines.
Results: Users can find ZebraShare submissions by searching ZFIN for specific alleles or line designations, just as for alleles submitted through the normal process. We present several potential examples of submission types to ZebraShare including a phenotypic mutants, mildly phenotypic, and early lethal mutants. Mutants for kdm1a show no apparent skeletal phenotype, and phf21aa mutants show only a mild skeletal phenotype, yet these genes have specific human disease relevance and therefore may be useful for further studies. The p120-catenin encoding gene, ctnnd1, was knocked out to investigate a potential role in brain development or function. The homozygous ctnnd1 mutant disintegrates during early somitogenesis and the heterozygote has localized defects, revealing vital roles in early development. Two snu13 genes were knocked out to investigate a role in muscle formation. The snu13a;snu13b double mutant has an early embryonic lethal phenotype, potentially related to a proposed role in the core splicing complex. In each example, the mutants submitted to ZebraShare display phenotypes that are not ideally suited to their originating lab's project directions but may be of great relevance to other researchers.
Conclusion: ZebraShare provides an opportunity for researchers to directly share information about mutant lines within ZFIN, which is widely used by the community as a central database of information about zebrafish lines. Submissions of alleles with a phenotypic or unexpected phenotypes is encouraged to promote collaborations, disseminate lines, reduce redundancy of effort and to promote efficient use of time and resources. We anticipate that as submissions to ZebraShare increase, they will help build an ultimately more complete picture of zebrafish genetics and development.
2020
Schredelseker T & Driever W. (2020). Conserved Genoarchitecture of the Basal Hypothalamus in Zebrafish Embryos. Front Neuroanat , 14, 3. PMID: 32116574 DOI.
Conserved Genoarchitecture of the Basal Hypothalamus in Zebrafish Embryos
Analyses of genoarchitecture recently stimulated substantial revisions of anatomical models for the developing hypothalamus in mammalian and other vertebrate systems. The prosomeric model proposes the hypothalamus to be derived from the secondary prosencephalon, and to consist of alar and basal regions. The basal hypothalamus can further be subdivided into tuberal and mamillary regions, each with distinct subregions. Albeit being a widely used model system for neurodevelopmental studies, no detailed genoarchitectural maps exist for the zebrafish (Danio rerio) hypothalamus. Here, we compare expression domains of zebrafish genes, including arxa, shha, otpa, isl1, lhx5, nkx2.1, nkx2.2a, pax6, and dlx5a, the orthologs of which delimit specific subregions within the murine basal hypothalamus. We develop the highly conserved brain-specific homeobox (bsx) gene as a novel marker for genoarchitectural analysis of hypothalamic regions. Our comparison of gene expression patterns reveals that the genoarchitecture of the basal hypothalamus in zebrafish embryos 48 hours post fertilization is highly similar to mouse embryos at E13.5. We found the tuberal hypothalamus in zebrafish embryos to be relatively large and to comprise previously ill-defined regions around the posterior hypothalamic recess. The mamillary hypothalamus is smaller and concentrates to rather medial areas in proximity to the anterior end of the neural tube floor plate. Within the basal hypothalamus we identified longitudinal and transverse tuberal and mamillary subregions topologically equivalent to those previously described in other vertebrates. However, the hypothalamic diencephalic boundary region and the posterior tuberculum still provide a challenge. We applied the updated prosomeric model to the developing zebrafish hypothalamus to facilitate cross-species comparisons. Accordingly, we applied the mammalian nomenclature of hypothalamic organization to zebrafish and propose it to replace some controversial previous nomenclature. Copyright © 2020 Schredelseker and Driever. KEYWORDS: bsx brain-specific homeobox; genoarchitecture; mamillary region; patterning; progenitor domains; prosomeric model; tuberal hypothalamus; zebrafish brain development PMCID: PMC7016197 DOI: 10.3389/fnana.2020.00003
Li J & Ge W. (2020). Zebrafish as a model for studying ovarian development: Recent advances from targeted gene knockout studies. Mol. Cell. Endocrinol. , 507, 110778. PMID: 32142861 DOI.
Zebrafish as a model for studying ovarian development: Recent advances from targeted gene knockout studies
Ovarian development is a complex process controlled by precise coordination of multiple factors. The targeted gene knockout technique is a powerful tool to study the functions of these factors. The successful application of this technique in mice in the past three decades has significantly enhanced our understanding on the molecular mechanism of ovarian development. Recently, with the advent of genome editing techniques, targeted gene knockout research can be carried out in many species. Zebrafish has emerged as an excellent model system to study the control of ovarian development. Dozens of genes related to ovarian development have been knocked out in zebrafish in recent years. Much new information and perspectives on the molecular mechanism of ovarian development have been obtained from these mutant zebrafish. Some findings have challenged conventional views. Several genes have been identified for the first time in vertebrates to control ovarian development. Focusing on ovarian development, the purpose of this review is to briefly summarize recent findings using these gene knockout zebrafish models, and compare these findings with mammalian models. These established mutants and rapid development of gene knockout techniques have prompted zebrafish as an ideal animal model for studying ovarian development. Copyright © 2020 The Author(s). Published by Elsevier B.V. All rights reserved. KEYWORDS: CRISPR/Cas9; Gene knockout; Germ cell; Ovary; TALEN; ZFN; Zebrafish DOI: 10.1016/j.mce.2020.110778
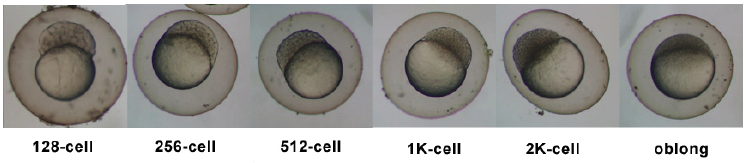
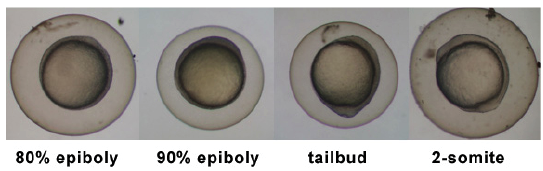
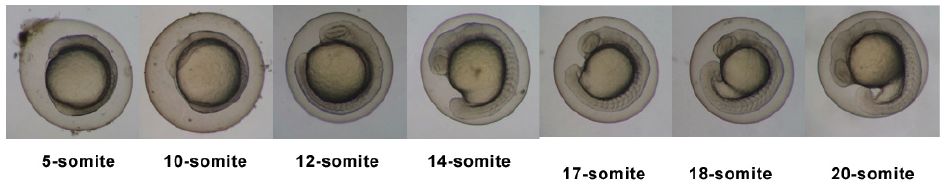
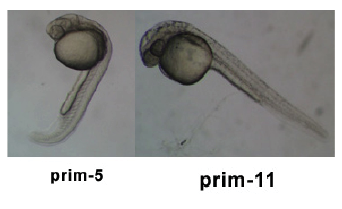
Stages
2019
Nagpal J, Herget U, Choi MK & Ryu S. (2019). Anatomy, development, and plasticity of the neurosecretory hypothalamus in zebrafish. Cell Tissue Res. , 375, 5-22. PMID: 30109407 DOI.
Anatomy, development, and plasticity of the neurosecretory hypothalamus in zebrafish
The paraventricular nucleus (PVN) of the hypothalamus harbors diverse neurosecretory cells with critical physiological roles for the homeostasis. Decades of research in rodents have provided a large amount of information on the anatomy, development, and function of this important hypothalamic nucleus. However, since the hypothalamus lies deep within the brain in mammals and is difficult to access, many questions regarding development and plasticity of this nucleus still remain. In particular, how different environmental conditions, including stress exposure, shape the development of this important nucleus has been difficult to address in animals that develop in utero. To address these open questions, the transparent larval zebrafish with its rapid external development and excellent genetic toolbox offers exciting opportunities. In this review, we summarize recent information on the anatomy and development of the neurosecretory preoptic area (NPO), which represents a similar structure to the mammalian PVN in zebrafish. We will then review recent studies on the development of different cell types in the neurosecretory hypothalamus both in mouse and in fish. Lastly, we discuss stress-induced plasticity of the PVN mainly discussing the data obtained in rodents, but pointing out tools and approaches available in zebrafish for future studies. This review serves as a primer for the currently available information relevant for studying the development and plasticity of this important brain region using zebrafish. KEYWORDS: Hypothalamus; Paraventricular nucleus; Stress; Zebrafish DOI: 10.1007/s00441-018-2900-4
Zebrafish znfl1s regulate left-right asymmetry patterning through controlling the expression of fgfr1a
Li J, Gao F, Zhao Y, He L, Huang Y, Yang X, Zhou Y, Yu L, Zhao Q & Dong X. (2019). Zebrafish znfl1s regulate left-right asymmetry patterning through controlling the expression of fgfr1a. J. Cell. Physiol. , 234, 1987-1995. PMID: 30317609 DOI.
J Cell Physiol. 2019 Mar;234(3):1987-1995. doi: 10.1002/jcp.27564. Epub 2018 Oct 14.
Li J1, Gao F2, Zhao Y2, He L3, Huang Y2, Yang X2, Zhou Y1, Yu L2, Zhao Q3, Dong X2.
Author information
Abstract
Proper left-right (LR) axis establishment is critical for organogenesis in vertebrates. Previously, we reported that zinc finger transcription factors zinc finger transcription factor 1 (znfl1s) are expressed in the tailbud and axial mesoderm in zebrafish. However, a role of znfl1s in LR axis development has not been demonstrated. Here, we discovered that the knockdown of znfl1s using morpholino (MO) in whole embryos or dorsal forerunner cells (DFCs) interrupted LR asymmetry and normal development of the heart, liver, and pancreas. Whole-embryo knockdown of znfl1s by MO or clustered regularly interspaced short palindromic repeat (CRISPR) interference (CRISPRi) resulted in the absent expression of nodal gene spaw and Nodal signaling-related genes lft1, lft2, and pitx2c in the left lateral plate mesoderm (LPM), and Spaw, Lft1, Lft2, and Pitx2c play important roles in LR axis development in zebrafish. However, specific knockdown of znfl1s in DFCs resulted in random expression of spaw, lft1, lft2, and pitx2c. Knockdown of znfl1s led to abnormal cilia formation by the downregulation of fgfr1a and foxj1a expression. The expression of spaw, lft1, lft2, and pitx2c was partially rescued by the overexpression of fgfr1a mRNA in znfl1s morphants. Taken together, our results suggest that znfl1s regulate laterality development in zebrafish embryos through controlling the expression of fgfr1a.
© 2018 Wiley Periodicals, Inc.
KEYWORDS:
Nodal-Pitx2 pathway; fgfr1a; foxj1a; left-right (LR) asymmetry; zebrafish; zinc finger transcription factor 1 (znfl1s)
PMID: 30317609 DOI: 10.1002/jcp.27564
2016
A crystal-clear zebrafish for in vivo imaging
Sci Rep. 2016 Jul 6;6:29490. doi: 10.1038/srep29490.
Antinucci P1, Hindges R1.
Abstract
The larval zebrafish (Danio rerio) is an excellent vertebrate model for in vivo imaging of biological phenomena at subcellular, cellular and systems levels. However, the optical accessibility of highly pigmented tissues, like the eyes, is limited even in this animal model. Typical strategies to improve the transparency of zebrafish larvae require the use of either highly toxic chemical compounds (e.g. 1-phenyl-2-thiourea, PTU) or pigmentation mutant strains (e.g. casper mutant). To date none of these strategies produce normally behaving larvae that are transparent in both the body and the eyes. Here we present crystal, an optically clear zebrafish mutant obtained by combining different viable mutations affecting skin pigmentation. Compared to the previously described combinatorial mutant casper, the crystal mutant lacks pigmentation also in the retinal pigment epithelium, therefore enabling optical access to the eyes. Unlike PTU-treated animals, crystal larvae are able to perform visually guided behaviours, such as the optomotor response, as efficiently as wild type larvae. To validate the in vivo application of crystal larvae, we performed whole-brain light-sheet imaging and two-photon calcium imaging of neural activity in the retina. In conclusion, this novel combinatorial pigmentation mutant represents an ideal vertebrate tool for completely unobstructed structural and functional in vivo investigations of biological processes, particularly when imaging tissues inside or between the eyes.
PMID 27381182
2015
There and back again: development and regeneration of the zebrafish lateral line system
Wiley Interdiscip Rev Dev Biol. 2015 Jan-Feb;4(1):1-16. doi: 10.1002/wdev.160. Epub 2014 Oct 20.
Thomas ED, Cruz IA, Hailey DW, Raible DW.
Abstract
The zebrafish lateral line is a sensory system used to detect changes in water flow. It is comprised of clusters of mechanosensory hair cells called neuromasts. The lateral line is initially established by a migratory group of cells, called a primordium, that deposits neuromasts at stereotyped locations along the surface of the fish. Wnt, FGF, and Notch signaling are all important regulators of various aspects of lateral line development, from primordium migration to hair cell specification. As zebrafish age, the organization of the lateral line becomes more complex in order to accommodate the fish's increased size. This expansion is regulated by many of the same factors involved in the initial development. Furthermore, unlike mammalian hair cells, lateral line hair cells have the capacity to regenerate after damage. New hair cells arise from the proliferation and differentiation of surrounding support cells, and the molecular and cellular pathways regulating this are beginning to be elucidated. All in all, the zebrafish lateral line has proven to be an excellent model in which to study a diverse array of processes, including collective cell migration, cell polarity, cell fate, and regeneration. © 2014 Wiley Periodicals, Inc. PMID 25330982
2014
Construction of a vertebrate embryo from two opposing morphogen gradients
Science. 2014 Apr 4;344(6179):87-9. doi: 10.1126/science.1248252.
Xu PF1, Houssin N, Ferri-Lagneau KF, Thisse B, Thisse C. Author information
Abstract
Development of vertebrate embryos involves tightly regulated molecular and cellular processes that progressively instruct proliferating embryonic cells about their identity and behavior. Whereas numerous gene activities have been found to be essential during early embryogenesis, little is known about the minimal conditions and factors that would be sufficient to instruct pluripotent cells to organize the embryo. Here, we show that opposing gradients of bone morphogenetic protein (BMP) and Nodal, two transforming growth factor family members that act as morphogens, are sufficient to induce molecular and cellular mechanisms required to organize, in vivo or in vitro, uncommitted cells of the zebrafish blastula animal pole into a well-developed embryo.
PMID 24700857
2013
FishFace: interactive atlas of zebrafish craniofacial development at cellular resolution
BMC Dev Biol. 2013 May 28;13:23. doi: 10.1186/1471-213X-13-23.
Eames BF, DeLaurier A, Ullmann B, Huycke TR, Nichols JT, Dowd J, McFadden M, Sasaki MM, Kimmel CB. Source Institute of Neuroscience, University of Oregon, Eugene, OR, USA. b.frank@usask.ca
Abstract
BACKGROUND: The vertebrate craniofacial skeleton may exhibit anatomical complexity and diversity, but its genesis and evolution can be understood through careful dissection of developmental programs at cellular resolution. Resources are lacking that include introductory overviews of skeletal anatomy coupled with descriptions of craniofacial development at cellular resolution. In addition to providing analytical guidelines for other studies, such an atlas would suggest cellular mechanisms underlying development. DESCRIPTION: We present the Fish Face Atlas, an online, 3D-interactive atlas of craniofacial development in the zebrafish Danio rerio. Alizarin red-stained skulls scanned by fluorescent optical projection tomography and segmented into individual elements provide a resource for understanding the 3D structure of the zebrafish craniofacial skeleton. These data provide the user an anatomical entry point to confocal images of Alizarin red-stained zebrafish with transgenically-labelled pharyngeal arch ectomesenchyme, chondrocytes, and osteoblasts, which illustrate the appearance, morphogenesis, and growth of the mandibular and hyoid cartilages and bones, as viewed in live, anesthetized zebrafish during embryonic and larval development. Confocal image stacks at high magnification during the same stages provide cellular detail and suggest developmental and evolutionary hypotheses. CONCLUSION: The FishFace Atlas is a novel learning tool for understanding craniofacial skeletal development, and can serve as a reference for a variety of studies, including comparative and mutational analyses.
FishFace Atlas https://www.facebase.org/fishface/home
Online zebrafish atlases include the Zebrafish Atlas (zfatlas.psu.edu); 3D Atlas of Zebrafish Vasculature Anatomy (http://uvo.nichd.nih.gov/atlas.html); the Zebrafish Brain Atlas (http://www.ucl.ac.uk/zebrafish-group/zebrafishbrain/index.php); the Atlas of Zebrafish Anatomy (http://www.zebrafish.uni-freiburg.de/anatomy.html); the Atlas of Zebrafish Development (http://bio-imaging.liacs.nl/liacsatlas.html); the Zebrafish Anatomy Portal (http://www.zfap.org); and the FishNet 3D developmental atlas (http://www.fishnet.org.au/index.shtml).
PMID 23714426
2012
Using the Tg(nrd:egfp)/albino zebrafish line to characterize in vivo expression of neurod
PLoS One. 2012;7(1):e29128. doi: 10.1371/journal.pone.0029128. Epub 2012 Jan 3.
Thomas JL, Ochocinska MJ, Hitchcock PF, Thummel R. Source Department of Anatomy and Cell Biology and Department of Ophthalmology, Wayne State University School of Medicine, Detroit, Michigan, United States of America.
Abstract
In this study, we used a newly-created transgenic zebrafish, Tg(nrd:egfp)/albino, to further characterize the expression of neurod in the developing and adult retina and to determine neurod expression during adult photoreceptor regeneration. We also provide observations regarding the expression of neurod in a variety of other tissues. In this line, EGFP is found in cells of the developing and adult retina, pineal gland, cerebellum, olfactory bulbs, midbrain, hindbrain, neural tube, lateral line, inner ear, pancreas, gut, and fin. Using immunohistochemistry and in situ hybridization, we compare the expression of the nrd:egfp transgene to that of endogenous neurod and to known retinal cell types. Consistent with previous data based on in situ hybridizations, we show that during retinal development, the nrd:egfp transgene is not expressed in proliferating retinal neuroepithelium, and is expressed in a subset of retinal neurons. In contrast to previous studies, nrd:egfp is gradually re-expressed in all rod photoreceptors. During photoreceptor regeneration in adult zebrafish, in situ hybridization reveals that neurod is not expressed in Müller glial-derived neuronal progenitors, but is expressed in photoreceptor progenitors as they migrate to the outer nuclear layer and differentiate into new rod photoreceptors. During photoreceptor regeneration, expression of the nrd:egfp matches that of neurod. We conclude that Tg(nrd:egfp)/albino is a good representation of endogenous neurod expression, is a useful tool to visualize neurod expression in a variety of tissues and will aid investigating the fundamental processes that govern photoreceptor regeneration in adults.
PMID 22235264
Manual drainage of the zebrafish embryonic brain ventricles
J Vis Exp. 2012 Dec 16;(70). pii: 4243. doi: 10.3791/4243.
Chang JT, Sive H. Source Department of Biology, Whitehead Institute of Biomedical Research, Massachusetts Institute of Technology.
Abstract
Cerebrospinal fluid (CSF) is a protein rich fluid contained within the brain ventricles. It is present during early vertebrate embryonic development and persists throughout life. Adult CSF is thought to cushion the brain, remove waste, and carry secreted molecules(1,2). In the adult and older embryo, the majority of CSF is made by the choroid plexus, a series of highly vascularized secretory regions located adjacent to the brain ventricles(3-5). In zebrafish, the choroid plexus is fully formed at 144 hours post fertilization (hpf)(6). Prior to this, in both zebrafish and other vertebrate embryos including mouse, a significant amount of embryonic CSF (eCSF) is present . These data and studies in chick suggest that the neuroepithelium is secretory early in development and may be the major source of eCSF prior to choroid plexus development(7). eCSF contains about three times more protein than adult CSF, suggesting that it may have an important role during development(8,9). Studies in chick and mouse demonstrate that secreted factors in the eCSF, fluid pressure, or a combination of these, are important for neurogenesis, gene expression, cell proliferation, and cell survival in the neuroepithelium(10-20). Proteomic analyses of human, rat, mouse, and chick eCSF have identified many proteins that may be necessary for CSF function. These include extracellular matrix components, apolipoproteins, osmotic pressure regulating proteins, and proteins involved in cell death and proliferation(21-24). However, the complex functions of the eCSF are largely unknown. We have developed a method for removing eCSF from zebrafish brain ventricles, thus allowing for identification of eCSF components and for analysis of the eCSF requirement during development. Although more eCSF can be collected from other vertebrate systems with larger embryos, eCSF can be collected from the earliest stages of zebrafish development, and under genetic or environmental conditions that lead to abnormal brain ventricle volume or morphology. Removal and collection of eCSF allows for mass spectrometric analysis, investigation of eCSF function, and reintroduction of select factors into the ventricles to assay their function. Thus the accessibility of the early zebrafish embryo allows for detailed analysis of eCSF function during development.
PMID 23271011
Vascular Endothelial Growth Factor Signaling Regulates the Segregation of Artery and Vein via ERK Activity during Vascular Development
Biochem Biophys Res Commun. 2012 Dec 21. pii: S0006-291X(12)02424-2. doi: 10.1016/j.bbrc.2012.12.076. [Epub ahead of print]
Kim SH, Schmitt CE, Woolls MJ, Holland MB, Kim JD, Jin SW. Source McAllister Heart Institute, and Curriculum in Genetics and Molecular Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
Abstract
Segregation of two axial vessels, the dorsal aorta and caudal vein, is one of the earliest patterning events occur during development of vasculature. Despite the importance of this process and recent advances in our understanding on vascular patterning during development, molecular mechanisms that coordinate the segregation of axial vessels remain largely elusive. In this report, we find that Vascular Endothelial Growth Factor-A (Vegf-A) signaling regulates the segregation of dorsal aorta and axial vein during development. Inhibition of Vegf-A pathway components including ligand Vegf-A and its cognate receptor Kdrl, caused failure in segregation of axial vessels in zebrafish embryos. Similarly, chemical inhibition of Mitogen-activated protein kinase kinase (Map2k1)/ Extracellular-signal-regulated kinases (Erk) and Phosphatidylinositol 3-kinases (PI3K), which are downstream effectors of Vegf-A signaling pathway, led to the fusion of two axial vessels. Moreover, we find that restoring Erk activity by over-expression of constitutively active MEK in embryos with a reduced level of Vegf-A signaling can rescue the defects in axial vessel segregation. Taken together, our data show that segregation of axial vessels requires the function of Vegf-A signaling, and Erk may function as the major downstream effector in this process. Copyright © 2012. Published by Elsevier Inc.
PMID 23266606
2011
Multifactorial Origins of Heart and Gut Defects in nipbl-Deficient Zebrafish, a Model of Cornelia de Lange Syndrome
Muto A, Calof AL, Lander AD, Schilling TF.Source Department of Developmental and Cell Biology, University of California, Irvine, California, United States of America.
Abstract
Cornelia de Lange Syndrome (CdLS) is the founding member of a class of multi-organ system birth defect syndromes termed cohesinopathies, named for the chromatin-associated protein complex cohesin, which mediates sister chromatid cohesion. Most cases of CdLS are caused by haploinsufficiency for Nipped-B-like (Nipbl), a highly conserved protein that facilitates cohesin loading. Consistent with recent evidence implicating cohesin and Nipbl in transcriptional regulation, both CdLS cell lines and tissues of Nipbl-deficient mice show changes in the expression of hundreds of genes. Nearly all such changes are modest, however-usually less than 1.5-fold-raising the intriguing possibility that, in CdLS, severe developmental defects result from the collective action of many otherwise innocuous perturbations. As a step toward testing this hypothesis, we developed a model of nipbl-deficiency in zebrafish, an organism in which we can quantitatively investigate the combinatorial effects of gene expression changes. After characterizing the structure and embryonic expression of the two zebrafish nipbl genes, we showed that morpholino knockdown of these genes produces a spectrum of specific heart and gut/visceral organ defects with similarities to those in CdLS. Analysis of nipbl morphants further revealed that, as early as gastrulation, expression of genes involved in endodermal differentiation (sox32, sox17, foxa2, and gata5) and left-right patterning (spaw, lefty2, and dnah9) is altered. Experimental manipulation of the levels of several such genes-using RNA injection or morpholino knockdown-implicated both additive and synergistic interactions in causing observed developmental defects. These findings support the view that birth defects in CdLS arise from collective effects of quantitative changes in gene expression. Interestingly, both the phenotypes and gene expression changes in nipbl morphants differed from those in mutants or morphants for genes encoding cohesin subunits, suggesting that the transcriptional functions of Nipbl cannot be ascribed simply to its role in cohesin loading.
PMID 22039349
2010
The zebrafish transcriptome during early development
BMC Dev Biol. 2011 May 24;11(1):30.
Vesterlund L, Jiao H, Unneberg P, Hovatta O, Kere J. Source Department of Biosciences and Nutrition, and Science for Life Laboratory, Karolinska Institutet, Stockholm, Sweden. liselotte.vesterlund@ki.se.
Abstract ABSTRACT: BACKGROUND: The transition from fertilized egg to embryo is accompanied by a multitude of changes in gene expression, and the transcriptional events that underlie these processes have not yet been fully characterized. In this study RNA-Seq is used to compare the transcription profiles of four early developmental stages in zebrafish (Danio rerio) on a global scale. RESULTS: An average of 79 M total reads were detected from the different stages. Out of the total number of reads 65% - 73% reads were successfully mapped and 36% - 44% out of those were uniquely mapped. The total number of detected unique gene transcripts was 11187, of which 10096 were present at 1-cell stage. The largest number of common transcripts was observed between 1-cell stage and 16-cell stage. An enrichment of gene transcripts with molecular functions of DNA binding, protein folding and processing as well as metal ion binding was observed with progression of development. The sequence data (accession number ERP000635) is available at the European Nucleotide Archive. CONCLUSION: Clustering of expression profiles shows that a majority of the detected gene transcripts are present at steady levels, and thus a minority of the gene transcripts clusters as increasing or decreasing in expression over the four investigated developmental stages. The three earliest developmental stages were similar when comparing highly expressed genes, whereas the 50% epiboly stage differed from the other three stages in the identity of highly expressed genes, number of uniquely expressed genes and enrichment of GO molecular functions. Taken together, these observations indicate a major transition in gene regulation and transcriptional activity taking place between the 512-cell and 50% epiboly stages, in accordance with previous studies.
PMID: 21609443
B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo
PLoS Genet. 2010 May 6;6:e1000936.
Okuda Y, Ogura E, Kondoh H, Kamachi Y.
Graduate School of Frontier Biosciences, Osaka University, Suita, Japan. Abstract The B1 SOX transcription factors SOX1/2/3/19 have been implicated in various processes of early embryogenesis. However, their regulatory functions in stages from the blastula to early neurula remain largely unknown, primarily because loss-of-function studies have not been informative to date. In our present study, we systematically knocked down the B1 sox genes in zebrafish. Only the quadruple knockdown of the four B1 sox genes sox2/3/19a/19b resulted in very severe developmental abnormalities, confirming that the B1 sox genes are functionally redundant. We characterized the sox2/3/19a/19b quadruple knockdown embryos in detail by examining the changes in gene expression through in situ hybridization, RT-PCR, and microarray analyses. Importantly, these phenotypic analyses revealed that the B1 SOX proteins regulate the following distinct processes: (1) early dorsoventral patterning by controlling bmp2b/7; (2) gastrulation movements via the regulation of pcdh18a/18b and wnt11, a non-canonical Wnt ligand gene; (3) neural differentiation by regulating the Hes-class bHLH gene her3 and the proneural-class bHLH genes neurog1 (positively) and ascl1a (negatively), and regional transcription factor genes, e.g., hesx1, zic1, and rx3; and (4) neural patterning by regulating signaling pathway genes, cyp26a1 in RA signaling, oep in Nodal signaling, shh, and mdkb. Chromatin immunoprecipitation analysis of the her3, hesx1, neurog1, pcdh18a, and cyp26a1 genes further suggests a direct regulation of these genes by B1 SOX. We also found an interesting overlap between the early phenotypes of the B1 sox quadruple knockdown embryos and the maternal-zygotic spg embryos that are devoid of pou5f1 activity. These findings indicate that the B1 SOX proteins control a wide range of developmental regulators in the early embryo through partnering in part with Pou5f1 and possibly with other factors, and suggest that the B1 sox functions are central to coordinating cell fate specification with patterning and morphogenetic processes occurring in the early embryo.
PMID: 20463883
The gastric mucosa development and differentiation
Prog Mol Biol Transl Sci. 2010;96:93-115.
Khurana S, Mills JC.
Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, Missouri, USA; Department of Developmental Biology, Washington University School of Medicine, St. Louis, Missouri, USA. Abstract The development and differentiation of the gastric mucosa are controlled by a complex interplay of signaling proteins and transcriptional regulators. This process is complicated by the fact that the stomach is derived from two germ layers, the endoderm and the mesoderm, with the first giving rise to the mature epithelium and the latter contributing the smooth muscle required for peristalsis. Reciprocal epithelial-mesenchymal interactions dictate the formation of the stomach during fetal development, and also contribute to its continuous regeneration and differentiation throughout adult life. In this chapter, we discuss the discoveries that have been made in different model systems, from zebrafish to human, which show that the Hedgehog, Wnt, Notch, bone morphogenetic protein, and fibroblast growth factor (FGF) signaling systems play essential roles during various stages of stomach development.
Copyright © 2010 Elsevier Inc. All rights reserved. PMID: 21075341
Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes
Carney TJ, Feitosa NM, Sonntag C, Slanchev K, Kluger J, Kiyozumi D, Gebauer JM, Coffin Talbot J, Kimmel CB, Sekiguchi K, Wagener R, Schwarz H, Ingham PW, Hammerschmidt M. PLoS Genet. 2010 Apr 15;6(4):e1000907. PMID: 20419147 [PubMed - indexed for MEDLINE]Free PMC ArticleFree text
Modes of developmental outgrowth and shaping of a craniofacial bone in zebrafish
Kimmel CB, DeLaurier A, Ullmann B, Dowd J, McFadden M. PLoS One. 2010 Mar 5;5(3):e9475.
PLoS One. 2010 Mar 5;5(3):e9475. Modes of developmental outgrowth and shaping of a craniofacial bone in zebrafish. Kimmel CB, DeLaurier A, Ullmann B, Dowd J, McFadden M.
Institute of Neuroscience, University of Oregon, Eugene, Oregon, United States of America. kimmel@uoneuro.uoregon.edu Abstract The morphologies of individual bones are crucial for their functions within the skeleton, and vary markedly during evolution. Recent studies have begun to reveal the detailed molecular genetic pathways that underlie skeletal morphogenesis. On the other hand, understanding of the process of morphogenesis itself has not kept pace with the molecular work. We examined, through an extended period of development in zebrafish, how a prominent craniofacial bone, the opercle (Op), attains its adult morphology. Using high-resolution confocal imaging of the vitally stained Op in live larvae, we show that the bone initially appears as a simple linear spicule, or spur, with a characteristic position and orientation, and lined by osteoblasts that we visualize by transgenic labeling. The Op then undergoes a stereotyped sequence of shape transitions, most notably during the larval period occurring through three weeks postfertilization. New shapes arise, and the bone grows in size, as a consequence of anisotropic addition of new mineralized bone matrix along specific regions of the pre-existing bone surfaces. We find that two modes of matrix addition, spurs and veils, are primarily associated with change in shape, whereas a third mode, incremental banding, largely accounts for growth in size. Furthermore, morphometric analyses show that shape development and growth follow different trajectories, suggesting separate control of bone shape and size. New osteoblast arrangements are associated with new patterns of matrix outgrowth, and we propose that fine developmental regulation of osteoblast position is a critical determinant of the spatiotemporal pattern of morphogenesis.
PMID 20221441
2009
Fishing for the genetic basis of cardiovascular disease
Tillman Dahme, Hugo A. Katus, and Wolfgang Rottbauer Dis Model Mech. 2009 Jan–Feb; 2(1-2): 18–22. doi: 10.1242/dmm.000687. PMCID: PMC2615162
<pubmed>19384962</pubmed>
"Excess retinoic acid (RA) signaling can be teratogenic and result in cardiac birth defects, but the cellular and molecular origins of these defects are not well understood. Excessive RA signaling can completely eliminate heart formation in the zebrafish embryo. However, atrial and ventricular cells are differentially sensitive to more modest increases in RA signaling. Increased Hox activity, downstream of RA signaling, causes phenotypes similar to those resulting from excess RA. These results suggest that Hox activity mediates the differential effects of ectopic RA on atrial and ventricular cardiomyocytes and may underlie the teratogenic effects of RA on the heart."
1981
Morphogenesis and synaptogenesis of the zebrafish Mauthner neuron
Kimmel CB, Sessions SK, Kimmel RJ. J Comp Neurol. 1981 May 1;198(1):101-20. PMID: 7229136