File:Hypothalamus gene interaction model.jpg
Original file (1,000 × 695 pixels, file size: 60 KB, MIME type: image/jpeg)
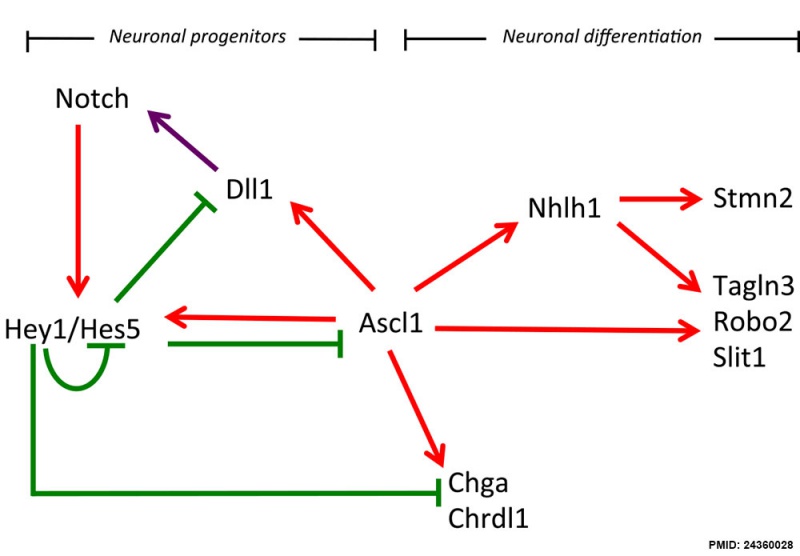
Hypothalamus Development Gene Interaction Model
Summary of potential regulatory interaction model of gene regulation for neurogenesis in the hypothalamus revealed by in silico analysis and literature.
A model of interaction between the negative regulators HES5 and HEY1 and the positive regulators ASCL1 and NHLH1 based on the literature and present data.
- Activations - identified with a red arrow
- Repressions - identified with a green barred line.
- Purple arrow - activation of NOTCH receptor by its ligand DLL1.
(Bertrand, 2002; Gohkle, 2008, Castro, 2011, de la Pompa 1997, and paper data).
- Links: Hypothalamus | Notch
Reference
Ratié L, Ware M, Barloy-Hubler F, Romé H, Gicquel I, Dubourg C, David V & Dupé V. (2013). Novel genes upregulated when NOTCH signalling is disrupted during hypothalamic development. Neural Dev , 8, 25. PMID: 24360028 DOI.
Copyright
© 2013 Ratié et al.; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
Ratié et al. Neural Development 2013 8:25 doi:10.1186/1749-8104-8-25
1749-8104-8-25-5.jpg
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 22:44, 23 March 2014 |  | 1,000 × 695 (60 KB) | Z8600021 (talk | contribs) | hypothalamus 1749-8104-8-25-5.jpg |
You cannot overwrite this file.
File usage
The following 3 pages use this file: