File:Proposed Hox protein classification.jpg
Original file (1,000 × 622 pixels, file size: 126 KB, MIME type: image/jpeg)
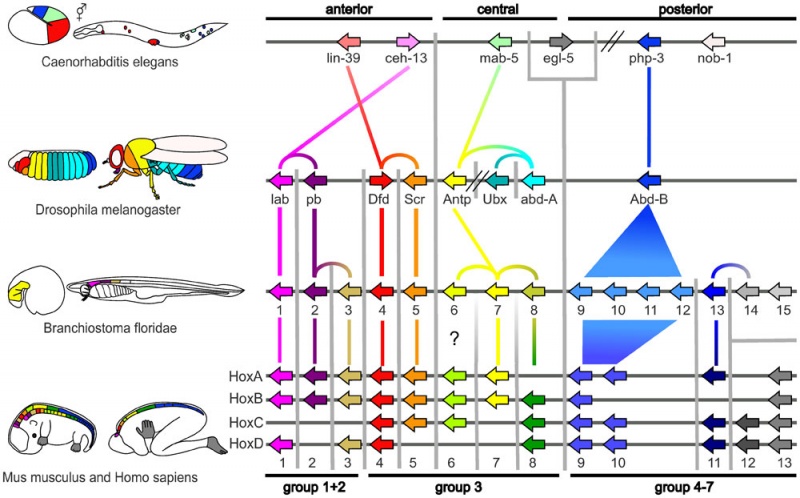
Proposed Hox Protein Classification
The organisms are ordered to show the clearest representation of the Hox-proteins most similar in sequence and thus expected to be most similar in function.
The figure is not supposed to indicate that Drosophila is descended from Caenorhabditis or that the chordates descended from Drosophila. Vertical gray lines delineate sequence similarity groups.
Colored lines linking Hox-genes indicate which Hox-proteins are most sequence similar to one another. Links within a species indicate a presumed multiplication, or loss, of the corresponding proteins in a lineage, while links between species indicate the most sequence similar pairs of Hox-proteins in these species.
The colors are used to represent groups of similar sequences, except for the ‘non-colors’ white and gray. These ‘non-colors’ indicate proteins with considerable sequence divergence to any other sequence in the model organisms we compare.
Zebrafish is not depicted as the assignment between tetrapods and zebrafish is clear.
- Hox Links: image - Human homeobox genes | image -Proposed Hox protein classification | Developmental Signals - Homeobox | Category:Hox
Reference
Hueber SD, Weiller GF, Djordjevic MA & Frickey T. (2010). Improving Hox protein classification across the major model organisms. PLoS ONE , 5, e10820. PMID: 20520839 DOI.
Copyright
© 2010 Hueber et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Hueber SD, Weiller GF, Djordjevic MA, Frickey T (2010) Improving Hox Protein Classification across the Major Model Organisms. PLoS ONE 5(5): e10820. doi:10.1371/journal.pone.0010820
Figure 10. Proposed classification. Journal.pone.0010820.g010.png
Cite this page: Hill, M.A. (2024, April 27) Embryology Proposed Hox protein classification.jpg. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/File:Proposed_Hox_protein_classification.jpg
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 18:06, 26 July 2010 |  | 1,000 × 622 (126 KB) | S8600021 (talk | contribs) | ==Proposed Hox Protein Classification== The organisms are ordered to show the clearest representation of the Hox-proteins most similar in sequence and thus expected to be most similar in function. The figure is not supposed to indicate that Drosophila |
You cannot overwrite this file.
File usage
The following 6 pages use this file: