File:Notch signaling pathway cartoon 01.jpg
Original file (1,211 × 1,280 pixels, file size: 235 KB, MIME type: image/jpeg)
Notch and its Signaling Pathway
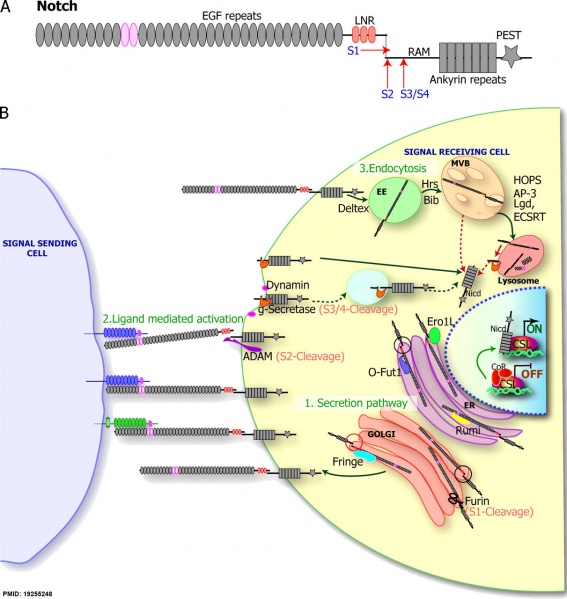
(A) The N-terminal part of the Notch ectodomain consists of 36 EGF-like repeats (gray ovals) and three LNRs (lin-12 Notch repeat; orange ovals). EGF repeats 11 and 12 interact with the ligands (pink ovals). NICD consists of an N-terminal RAM (recombination binding protein-J -associated molecule) domain, an ankyrin (ANK) domain (gray rectangles), and less-conserved regions, including a variable transactivation domain and a C-terminal PEST sequence (gray star). The red arrows indicate the cleavage sites: S1 (Furin), S2 (ADAM metalloprotease), and S3/S4 (γ-secretase).
(B, 1) Secretory pathway: the modifications during the secretion of Notch to the membrane in the ER (purple) and the Golgi (orange). Notch is translated inside ER, where it is glycosylated by an O-fucosyltransferase O-Fut1 (light purple) and O-glucosyltransferase Rumi (yellow). Note the black circle on the Notch molecule in the ER; because Notch is not cleaved, the extracellular and intracellular domains are physically linked. Notch is then translocated into Golgi, where it is cleaved by Furin protease (scissors) at the S1 site and further modified by the N-acetylglucosaminyltransferase, Fringe. Note the red circle on the Notch molecule in the Golgi after S1 cleavage; the extracellular and intracellular domains are not physically linked. (2) Ligand-mediated activation: Notch (gray) interacts with the DSL ligands, Delta (blue) and Serrate (green), resulting in a series of proteolytic cleavage events induced by ligand binding. The S2 cleavage is mediated by ADAM protease (purple), whereas the S3/4 cleavage event is mediated by γ-secretase (g-secretase, in orange). Several studies also suggest that γ-secretase–mediated cleavage can occur inside endocytic compartment (shown in the light blue circle). (3) The endocytic regulation of the Notch receptor: full-length Notch can undergo endocytosis, leading to translocation of Notch into EEs, MVB, and lysosomes. From genetic data, several proteins have been identified to modulate this process, including Hrs and Bib, possibly during the EE-to-MVB transition of Notch, Lgd, and ESCRT complex, or during the MVB-to-lysosomes transition. These proteins further modulate Notch activity as described in the text. The dotted red arrow shows that in mutants that affect trafficking from the MVB to the lysosome, or if Notch is not trafficked to the lumen of the lysosome, Notch can undergo γ-secretase cleavage, resulting in a Notch GOF phenotypes.
- Notch Links: Notch structure cartoon | Notch signaling pathway cartoon | Notch and signaling pathway cartoon | Developmental Signals - Notch | Molecular Factors
Reference
<pubmed>19255248</pubmed>
Copyright
Rockefeller University Press - Copyright Policy This article is distributed under the terms of an Attribution–Noncommercial–Share Alike–No Mirror Sites license for the first six months after the publication date (see http://www.jcb.org/misc/terms.shtml). After six months it is available under a Creative Commons License (Attribution–Noncommercial–Share Alike 4.0 Unported license, as described at https://creativecommons.org/licenses/by-nc-sa/4.0/ ). (More? Help:Copyright Tutorial)
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 20:46, 4 February 2015 |  | 1,211 × 1,280 (235 KB) | Z8600021 (talk | contribs) | ==Notch and its Signaling Pathway== (A) The N-terminal part of the Notch ectodomain consists of 36 EGF-like repeats (gray ovals) and three LNRs (lin-12 Notch repeat; orange ovals). EGF repeats 11 and 12 interact with the ligands (pink ovals). NICD con... |
You cannot overwrite this file.
File usage
There are no pages that use this file.