File:Blastomere genomic divergence 01.jpg
Blastomere_genomic_divergence_01.jpg (709 × 542 pixels, file size: 60 KB, MIME type: image/jpeg)
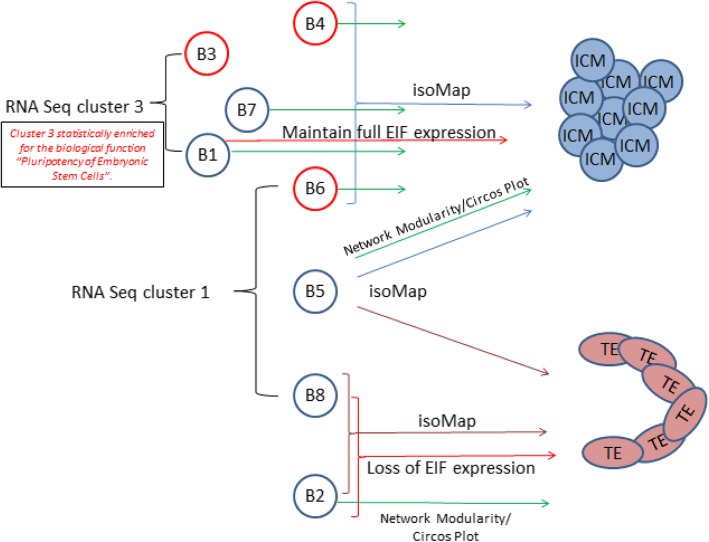
Summary schematic representing 8-cell blastomere genomic divergence
Using a combination of systems biology techniques including network analysis, nearest neighbour PCA (isoMap), EIF expression and epigenetic-associated gene expression we have detected two sets of blastomeres within an 8-cell embryo which exhibit diverging tendencies towards ICM and TE.
Nearest neighbour PCA analysis (isoMap) reveals similarities in global gene expression between blastomere 3, 4, 7, 1, 6 and 5 with Inner Cell Mass (Blue brackets/arrows), and blastomere 8 and 2 with Trophectoderm (burgundy brackets/arrows). Green arrows represent similarity in network modules, as highlighted by circos plots.
Black bracketed Blastomere 3, 7 and 8 are enriched in RNAseq cluster 3 and Blastomere 6, 5 and 8 are enriched in RNAseq cluster 1. Red brackets/arrows represent blastomeres with the greatest maintenance and loss of developmental EIF expression. B3, B4 and B6 (red coloured blastomeres) have distinct epigenetic associated gene expression signatures from those expressed by blastomeres B1, B2, B5, B7 and B8 (blue blastomeres)
Above text from figure legend.
- Links: blastocyst | trophoblast
Reference
Smith HL, Stevens A, Minogue B, Sneddon S, Shaw L, Wood L, Adeniyi T, Xiao H, Lio P, Kimber SJ & Brison DR. (2019). Systems based analysis of human embryos and gene networks involved in cell lineage allocation. BMC Genomics , 20, 171. PMID: 30836937 DOI.
Copyright
© The Author(s). 2019 Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6399968/figure/Fig7/?report=objectonly 12864_2019_5558_Fig7_HTML.jpg
Cite this page: Hill, M.A. (2024, April 27) Embryology Blastomere genomic divergence 01.jpg. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/File:Blastomere_genomic_divergence_01.jpg
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 05:57, 2 January 2020 |  | 709 × 542 (60 KB) | Z8600021 (talk | contribs) | ==Summary schematic representing 8-cell blastomere genomic divergence== Using a combination of systems biology techniques including network analysis, nearest neighbour PCA (isoMap), EIF expression and epigenetic-associated gene expression we have detected two sets of blastomeres within an 8-cell embryo which exhibit diverging tendencies towards ICM and TE. Nearest neighbour PCA analysis (isoMap) reveals similarities in global gene expression between blastomere 3, 4, 7, 1, 6 and 5 with Inner... |
You cannot overwrite this file.
File usage
The following page uses this file: