BGDB Sexual Differentiation - Sex Determination: Difference between revisions
No edit summary |
No edit summary |
||
| Line 49: | Line 49: | ||
* This population of cells then lie at the hindgut and yolk sac junctional region and later migrate into the germinal ridge in early embryonic development. | * This population of cells then lie at the hindgut and yolk sac junctional region and later migrate into the germinal ridge in early embryonic development. | ||
* It is not the primordial germ cells which respond to SRY presence or absence, but the supporting cells within the developing gonad. | * It is not the primordial germ cells which respond to SRY presence or absence, but the supporting cells within the developing gonad. | ||
{{BGDB SexDiffn}} | |||
==References== | ==References== | ||
Revision as of 12:15, 29 May 2011
| Practical 12: Sex Determination | Early Embryo | Late Embryo | Fetal | Postnatal | Abnormalities | 2011 Audio |
Introduction
Sex determination (male/female) at the biological level is determined by the presence or absence of the Y chromosome.
Initially, we did not know what this factor was and it was designated the "testis determining factor" (TDF). We now know (since 1990) that TDF is the protein product encoded by the SRY gene on the Y chromosome. Without this gene/protein the potential sex is female (see Male below).
For some time, female was considered the "default" sex in the absence of SRY, we now know this is not the case, with several genes specifically required for ovary formation. In females, sex determination involves at least one X chromosome gene, DAX1 encoding a nuclear hormone receptor.
Another critical genetic issue is related to the presence of two X chromosomes, "gene dosage", and in the case of mammals this is regulated by inactivating one of those X chromosomes in each and every cell (see Female below).
Male (XY)
Sry was discovered (1990) by studying a human XY female, resulting from a deletion in the Y chromosome that did not allow testis development. Subsequent mapping of this deletion allowed isolation and characterization of the SRY gene.
There is a suggestion that SRY may allow testes development by acting to inhibit DAX1, which is expressed in the indifferent gonad at the same time. The mechanism of an inhibitor inhibiting and inhibitor is seen in some other developing systems.
- encodes a 204 amino acid protein (Mr 23884 Da) that is a zinc-finger transcription factor.
- transcription factors bind to specific sites of DNA and regulates the transcription (expression) of other genes, we still do not know all the genes SRY regulates.
- expressed when testes begin to form, in gonadal tissue and does not require the presence of germ cells.
Notes: The Y chromosome is much smaller than the X chromosome and by definition, cannot contain important genes for other cellular functions.
Nomenclature, capital letters are used for human genes (SRY) and lower case letters are used for the equivalent genes in other species (sry).
Female (XX)
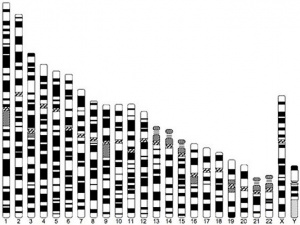
- In contrast to the Y chromosome, the X chromosome contains about 5% of the haploid genome and encodes house-keeping and specialized functions.
- The genetic content of the X chromosome has been strongly conserved between species.
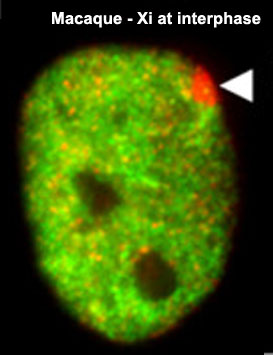
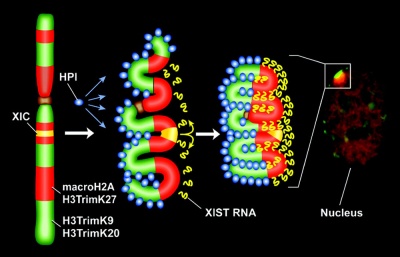
1961 - In order to have correct levels of X chromosome gene/protein expression (gene dosage), females must "inactivate" a single copy of the X chromosome (Xi) in each and every cell.
1991 - The initiator of the X inactivation process was discovered (1991) to be regulated by a region on the inactivating X chromosome encoding an X inactive specific transcript (XIST), that acts as RNA and does not encode a protein. Furthermore X inactivation occurs randomly throughout the embryo, generating a mosaic of maternal and paternally derived X chromosome activity in all tissues and organs.
- Links: Signaling in genital development | Fig. 1 - image | X Chromosome Inactivation - Epigenetics 1
Primordial Germ Cells
- Primordial Germ Cells (PGCs) are thought to be the first population of cells to migrate through the primitive streak in early gastrulation.
- This population of cells then lie at the hindgut and yolk sac junctional region and later migrate into the germinal ridge in early embryonic development.
- It is not the primordial germ cells which respond to SRY presence or absence, but the supporting cells within the developing gonad.
| Practical 12: Sex Determination | Early Embryo | Late Embryo | Fetal | Postnatal | Abnormalities | 2011 Audio |
References
BGDB: Lecture - Gastrointestinal System | Practical - Gastrointestinal System | Lecture - Face and Ear | Practical - Face and Ear | Lecture - Endocrine | Lecture - Sexual Differentiation | Practical - Sexual Differentiation | Tutorial
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, May 2) Embryology BGDB Sexual Differentiation - Sex Determination. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/BGDB_Sexual_Differentiation_-_Sex_Determination
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G