File:Hox deuterostomes phylogenetic tree.jpg: Difference between revisions
From Embryology
(==Phylogenetic tree of deuterostomes showing the Hox clusters of non-olfactores deuterostomes== The Hox repertoire of a substantial number of groups within the invertebrate deuterostomes is still lacking (black question marks), and the origin of a 15 ...) |
mNo edit summary |
||
| Line 3: | Line 3: | ||
The Hox repertoire of a substantial number of groups within the invertebrate deuterostomes is still lacking (black question marks), and the origin of a 15 Hox gene cluster in cephalochordates, or when the Hox4 was lost in echinoderms are still a mystery (indicated by red question marks and arrows). Yellow, anterior Hox genes; orange, Hox3; blue, central Hox genes; green, posterior Hox genes. | The Hox repertoire of a substantial number of groups within the invertebrate deuterostomes is still lacking (black question marks), and the origin of a 15 Hox gene cluster in cephalochordates, or when the Hox4 was lost in echinoderms are still a mystery (indicated by red question marks and arrows). Yellow, anterior Hox genes; orange, Hox3; blue, central Hox genes; green, posterior Hox genes. | ||
===Reference=== | |||
<pubmed>23819519</pubmed> | |||
1471-213X-13-26-1 | 1471-213X-13-26-1 | ||
| Line 8: | Line 12: | ||
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3707753/figure/F1/ | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3707753/figure/F1/ | ||
[[Category:Molecular]] [[Category:Hox]] [[Category:Cartoon]] | |||
Revision as of 18:05, 21 November 2013
Phylogenetic tree of deuterostomes showing the Hox clusters of non-olfactores deuterostomes
The Hox repertoire of a substantial number of groups within the invertebrate deuterostomes is still lacking (black question marks), and the origin of a 15 Hox gene cluster in cephalochordates, or when the Hox4 was lost in echinoderms are still a mystery (indicated by red question marks and arrows). Yellow, anterior Hox genes; orange, Hox3; blue, central Hox genes; green, posterior Hox genes.
Reference
<pubmed>23819519</pubmed>
1471-213X-13-26-1
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3707753/figure/F1/
File history
Yi efo/eka'e gwa ebo wo le nyangagi wuncin ye kamina wunga tinya nan
| Gwalagizhi | Nyangagi | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 18:04, 21 November 2013 | 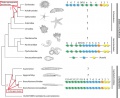 | 977 × 800 (108 KB) | Z8600021 (talk | contribs) | ==Phylogenetic tree of deuterostomes showing the Hox clusters of non-olfactores deuterostomes== The Hox repertoire of a substantial number of groups within the invertebrate deuterostomes is still lacking (black question marks), and the origin of a 15 ... |
You cannot overwrite this file.
File usage
The following page uses this file: