2009 Group Project 4
The Mouse (Mus musculus)
Introduction
The mouse is a small animal which can be used as a model to study embryological development. It belongs to the class of mammalian and order of rodentia and shares a significant similarity in homology with humans. The mouse is one of the most commonly used animals in experimental embryology and most sciences. The mouse is a useful mammalian model for human embryological development because the mouse:
- Has a very similar size genome as the human genome
- Has genes which can be easily manipulated and studied
- Has a high degree of homogeny with humans
- Produces a large offspring in a short amount of time
- Has no ethical issues limiting its use in experiments
- Is a small organism which can be easily maintained
- Is not expensive
History of the use of the Mouse Embryo Model
Introduction
The mouse has had a significant contribution to the field of Biomedicine since the middle of the sixteenth century (Hendrich et al. 2004). During the twentieth century, the use of the mouse embryo in particular, has increased significantly, and continues to be a popular experimental choice for researchers.
It's popularity over time in mammalian biology, biomedicine, immunology, oncology, pathology, and genetics is a result of:
- The small size of the mouse and its resistance to infection (Nagy et.al. 2003).
- The efficient mouse breeding system: Which consistently monitors the characterisitcs, that are precisley known, generation after generation, thereby producing highly standardised strains (Hedrich et al. 2004).
- The metabolic and internal anatomical similarities between the mouse and the human, which allows for comparisons. Because of these similarities, they share similar diseases, for example, cancer, diabetes, autoimmunity, endocrine disease, and neurological dysfunctions (Hedrich et al. 2004).
- The ability to manipulate the mouse germ line. This can be achieved by the:
1. Genetic manipulation of embryonic stem (ES) cells, or
2. Direct injection of cloned DNA into zygotes. See Genetics section below.
Please note that the following findings are based on research which have utilised the model of the mouse embryo.
Gregor Johann Mendel (1822-1884), Augustinian priest and scientist
In his notes, One Hundred Years of Mouse Genetics: An Intellectual History I The classical period (1902-1980), Kenneth Paigen wrote that Mendel's first experiment on the transmission of inheritance was made using mice segregating for coat colour markers. Due to complaints from members of the Catholic Church about the 'smell' of the mice , he changed his material to Pea plants (Hedrich et.al. 2004).
1895: Robert Heinrich Johannes Sobotta (1869-1945), German Anatomist [1]
What did he do? Researched the fertilisation and cleavage of the mouse's egg (Andrews, 1895). Link to Dr. Sobotta's research [2]
What did he find?
- The corpus Luteum is formed by the enlargement of the epithelial cells of the follicle, aided by growth of connective tissue.
- There is no distinction between corpora lutea vera and corpora lutea spuria.
- The corpora lutea do not degenerate. They remain unchanged during the life of the animal, and therefore add to the size of the ovary.
1949: John H. Hammond, Jr. (1888-1965) [3], Animal Husbandry Scientist
What did he do?
Cultured eight-cell morulae and four-cell-stage mouse embryos to the Blastocyst sage.
This was the first report of successful attempts to culture mouse embryos in vitro to the Blastocyst stage (Nagy et.al. 2003).
The embryos removed at the two-cell stage died a while after (Nagy et.al. 2003).
1956: Wesley Kingston Whitten (1918-), Australian Veterinary Scientist
Link to photo of Whesley Whitten: [4]
What did he do?
Cultured 8-cell mouse embryos to the Blastocyst stage using a medium containing Krebs-Ringer's bicarbonate solution supplemented with glucose and bovine serum albumin (Nagy et. el. 2003).
Soon after, he found that some two-cell-stage embryos developed into blastocysts, upon some modifications to the original medium.
1958: Dame Anne Laura Dorinthea McLaren (1927-2007), Developmental Biologist
Link to photo of Anne McLaren: [5]
What did she do? Developed the first birth of mice in-vitro. Link to Published article [6]
1962: George Todaro (1937-) and Howard Green, Cell Biologists
What did they do? They studied the fibroblast cells of the mouse embryo in culture.
What did they find?
- A few weeks after the culture, the growth rate of the Fibroblast cells slowed down. They thought that the cells, as with normal human fibroblast cells, would eventually stop dividing and die. However this was not the case.
- Two to three months later, the cell growth rate increased. This meant that the resulting new population of cells had undergone spontaneous transformation. The resulting permanently dividing cell line, called the '3T3 cell line', have been growing in culture for over 20 years. (Bhamrah et al. 2002)
- As a result, they explained how cells behave in a similar fashion once they have undergone transformation (Bhamrah et al. 2002).
1963: Ralph L. Brinster (1932-)[7], American Geneticist
What did he find?
- Determined the nutritional requirements of the pre-implantation mouse embryo.
- Established the microdrop technique that allowed two-cell-stage mouse embryos to be cultured to the Blastocyst stage (Nagy et.al. 2003).
1965: Beatrice Mintz (1921-)[8], American Embryologist
Link to photo of Beatrice Mintz: [9]
What did she do? Generated adult chimeras.
What did she find? The zona pelucida of the mouse embryo could be digested using pronase (Nagy et.al 2003).
1974: Rudolf Jaenisch (1942-), German biologist and Beatrice Mintz
What did they do? Injected purified SV40 (Simian vacuolating virus 40 or Simian virus 40 [10]) DNA into the mouse blastocyst cavity.
What did they find?
- Viral DNA sequences were detected in the somatic tissue of these mice. Therefore, the DNA had integrated into the genome of embryonic cells (Nagy et.al. 2003).
1974: Ralph Brinster [11], & 1975: Mintz and Illmensee [12], and Papaioannou et al. [13]
What did they find? Embryonic carcinoma (EC) stem cells could be injected into blastocysts to create adult chimeras that contained normal tissues derived from the EC cells.
1976: Rudolf Jaenisch
What did he do? Infected a preimplanted mouse embryo with the Moloney murine leukemia virus. By doing this, he introduced the virus into the mouse germ line in a stable manner.
The significance of this is outlined below under the findings of Anderson (1980), Capecchi (1980), and Gordon (1980). Overall, Jaenisch's work on the mouse embryo led to the findings of these mentioned researchers.
1977: R.J. Mullen
What did he do? Fused the normal and reeler mouse embryo at the morula stage to produce allophenic mice. (Rouvroit & Goffinet 1998)
1980: Ralph L. Brinster
What did he do? The first to inject purified globin mRNA into mouse zygotes (Nagy et.al. 2003).
This formed the basis for the production of transgenic mice.
1980: Wayne F. Anderson, Lydia Killos, L Sanders-Haigh, P J Kretschmer, and E G Diacumakos & 1980: Mario R. Capecchi
What did they do? Microinjected cloned herpes simplex virus (HSV) thymidine kinase (tk) gene into the nuclei of cultured TK-deficient mouse fibroblast cells.
What did they find? The incorporation and expression of the tk gene may be accomplished in a 'stable' manner.
This suggested that the microinjection of DNA into the one-cell mouse embryo might allow for the efficient introduction of cloned genes into the developing mouse (Nagy et.al. 2003). This triggered researchers, including Gordon (mentioned below), to test this possibility.
Click here for Anderson's published article, or click here for Capecchi's published article.
1980: J.W. Gordon, G A Scangos, D J Plotkin, J A Barbosa, and F H Ruddle
What did they do?
Directly injected the cloned HSV thymidine kinase (tk) gene into the pronuclei of zygotes, therefore, introducing the gene into the somatic tissues (Nagy et.al. 2003).
Click here for Gordon's published article.
1981: M.H. Kaufman & M.J. Evans, and 1981: G.R. Martin
What did they do? They were the first to develop ES cells from cultured mouse blastocysts (Nagy et.al. 2003).
Click here for Martin's published article.
1983: Davor Solter (1939-), Developmental Biologist
What did he do? Transferred nuclei between zygotes.
This revealed the importance of parent gene imprinting in mammalian development (Nagy et.al. 2003).
1986: Elizabeth Robertson, Allan Bradley, Michael Kuehn & Martin Evans
What did they do? They were the first to genetically manipulate ES cells (Nagy et.al. 2003).
This allows for the modification and selection of cells with germ-line potential. In turn, this allows them to derive the transgenic strains with pre-determined genetic changes.
By doing this, they inserted many proviral vector sequences that
- Provide new chromosomal molecular markers for linkage studies in the mouse, and
- May cause mutations [14].
1996: Toshio Ohshima, Jerrold M. Wardt, Chang-Goo Huht, Glenn Longenecker, Veeranna, Harish C.Pant, Roscoe O. Bradyt, Lee J. Martins, and Ashok B. Kulkarni
What did they do? Generated Cyclin-dependant kinase 5 (Cdk5) null mice, a type of mutant mice, by homologous recombination to assess the role of Cdk5 in vivo.
Targeted ES cells were injected into blastocysts to generate chimeric mice.
1998: Teruhiko Wakayama (1967-), Developmental Biologist and Ryuzo Yanagimachi (1928-), Animal Embryologist
Link to photo of Teruhiko Wakayama: [15]
What did they do? Cloned the first mouse, named Cumulina.
1998: Nikola Skreb and colleagues
What did they find? The early embryonic ectoderm contains cells capable of contributing to all three germ layers of the mouse fetus (Nagy et.al. 2003.
Use of teratocarcinomas
What are teratocarcinomas? They are '...gonadal tumors that contain a...mixture of different tissue types, all derived from a population of undifferentiated stem cells known as embryonal carcinoma cells.' (Nagy et.al. 2003)
The use of teratocarcinomas in research complement the studies on pluripotentiality of cells from the normal embryo and allows for further study of early embryonic development.
First pioneered by Leroy Stevens (1920-), Mammalian Embryologist & Barry Pierce
Link to photo of Leroy Stevens: [16]
Leroy C. Stevens, Jr.:
- The first to identify that male mice of the inbred 129 strain have a low incidence of testicular teratoma arising from primordial germ cells (Link to Stevens and C. C. Little's related article [17]).
- The first to identify modifier genes such as ter that increase the frequency of teratomas in the testis and eventually developed an inbred strain (129/Sv).
- Developed the LT strain, in which about 50% of females develop ovarian teratocarcinomas.
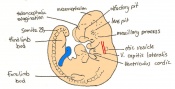
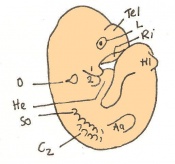
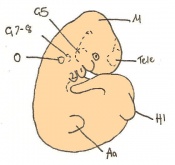
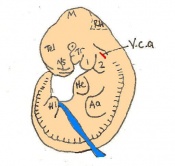
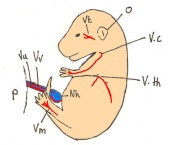
Staging Of Embryonic Development
The mouse model has been used significantly in past scientific experiments. The mouse has many beneficial qualities which allow it to be easily manipulate and studied. One example includes its ability to reproduce and embryological development.
Reproduction
Mice can reproduce all year round and are also polyestrous animals, meaning they can breed many times during the year. Male and female mice are capable of breeding at 50 days old and female mice begin their estrous cycle (go on 'heat') between 25-40 days old. The estrous cycle duration is 4-5 days and ovulation occurs spontaneously. A mouse produces an average litter of about 10-12 mice and begins a normal estrous cycle 3-10 days following partuition. Having an understanding of mouse reproduction, can highlight the useful nature of studying the mouse as a mamallian model for human emryology (Greenwald, 1956).
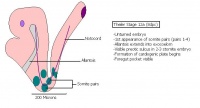
Theiler stages
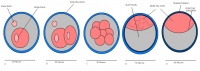
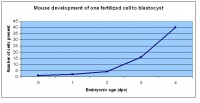
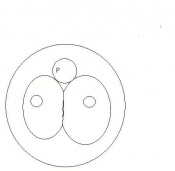
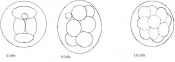
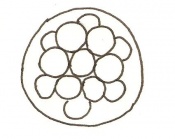
Mouse embryonic development commences once the female's egg or oocyte has become fertilized by the male's sperm. Mouse development has a gestation period of 19-21 days, and can range in different strains of mice. The development of an embryo can be categorized into different stages including cell number, somite stages and morphology. The most common method of staging is by Theiler (1989) which categorizes mouse development into prenatal and postnatal stages consisting of 26 and 2 stages respectively. Downs and Davies (1993) have also established a method of staging the mouse development based on morphological changes. Link to Downs and Davies[18] The developmental stages of the mouse embryo can be summarised in the tables and corresponding figures below in relation to characteristics at that stage. The first stage begins with fertiliation of the egg which divides into multiple cells to form a morulla and further go on to form a blastocyst which is ready for implantation (Nagy, et al., 2003).
| Theiler Stage | Embryonic Age in Days Post Coitum (dpc) | Stage Characteristic | Cell Characteristics | Zona Pellucida | Location | |
|---|---|---|---|---|---|---|
| 1 | 0-0.9
(range 0-0.25) |
One-celled embryo (fertilized) | One cell | Present | Ampulla | |
| 2 | 1
(range 1-2.5) |
Dividing egg | 2-4 cells
- 1st cleavage after 24hrs |
Present | Travelling down oviduct | |
| 3 | 2
(range 1-3.5) |
Morula (early to fully compacted) | 4-16 cells | Present | Oviduct (utero-tubal junction) | |
| 4 | 3
(range 2-4) |
Morula to Blastocyst
-Intra-cellular matrix -Blastocoelic cavity |
16-40 compacted cells
-Inner cell mass -Outer layer of trophectoderm cells |
Present | Uterine lumen | |
| 5 | 4
(range 3-5.5) |
Zona free Blastocyst (hatching) | Blastocyst implants as zona pelludica is lost | Absent | Uterine lumen |
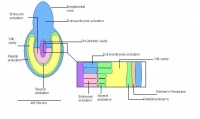
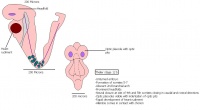
Once the Blastocyst has lost the zona pellucida, it is free to implant onto the uterine wall. After implantation multiple features of a trilaminar embryo start to develop. Ectoderm, endoderm and mesoderm layers form, which further develop many structures corresponding to different stages as summariesed in the table below. The developing mouse embryo which is implanted on the uterine wall can be viewed by ultrasonic radiation. Link to ultrasound of mouse embryo [19]
| Theiler Stage | Embryonic age in Days Post Coitum (dpc) | Stage Characteristic | Cell characteristics | |
|---|---|---|---|---|
| 6 | 4.5 (range 4-5.5)
Human carnegie stage: 4 |
Attachment of blastocyst
-Implantation |
Embryonic Endoderm | |
| 7 | 5 (range 4.5-6)
Human carnegie stage: 5 |
Implantation
-Egg cylinder -Ectoplacental cone |
Inner cell mass increases
-Epiblast -Mural trophectoderm lined by endoderm | |
| 8 | 6 (range 5-6.5)
Human carnegie stage: 5 |
embryonic and extra-embryonic regions
-Pro-amniotic cavity |
Trophoblast giant cells invade
-Maternal blood invades -Reichert's membrane | |
| 9 a) | Pre-streak | Advanced Endometrial and egg cylinder
-Embryonic axis |
Embryonic and extra-embryonic ectoderm
-Uterine crypts lose lumen | |
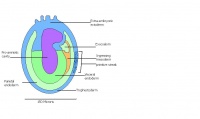
| 9 b) | Early streak | Gastrulation begins | Mesodermal cells | |
| 10 a) | 7 (range 6.5-7.5)
Mid to late streak Human carnegie stage: 8 |
Amnion formation | Amniotic fold
-Allantoic bud -Primitive node -Amnion closes | |
| 11 | 7.5 (range 7.25-8)
Human carnegie stage: 9 |
Neural plate and presomites | Amniotic cavity, exocoelom and ectoplacental cleft
-Allantoic bud elongates -Notochodal plate -Early head fold -Foregut pocket |
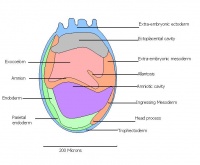
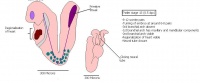
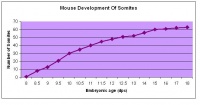
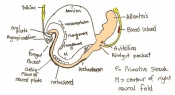
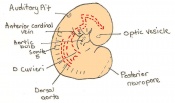
The next stage (Theiler stage 12) is marked by the first somite pair formation. Stages 13 onwards is characterized by increasing somite pairs up to 20 somite pairs. These embryo stages have characteristic features of developing forgut, prominent head fold, optic pits, branchial arch and neuropore formation. These four stages can be shown in table 3 below.
| Theiler stage | Embryonic age in Days Post Coitum (dpc) | Stage characteristic | Cell characteristics | Number of somite pairs | |
|---|---|---|---|---|---|
| 12 a) | 8 (range 7.5-8.75)
Human carnegie stage: 9 |
Unturned embryo
-1st somite pairs |
Allantois extends into exocoelom
-Maxillary components of 1st brachial arch prominent -Preotic sulcus -Cardiogenic plate -Foregut pocket |
1-4 | |
| 12 b) | 8 (range 7.5-8.75) | Unturned embryo | Prominent headfolds
-Neural closure -Optic placodes with optic pits -Rapid development of heart rudiment |
5-7 | |
| 13 | 8.5 (range 8-9.25)
Human carnegie stage: 10 |
Turning of embryo at around 6-8 pairs | 1st branchial arch with maxillary and mandibular components
-2nd branchial arch visible - Regionalization of heart visible -neural tube closure |
8-12 | |
| 14 | 9 (range 8.5-9.75)
Human carnegie stage: 11 |
Anterior neuropore formation and closure | optic pit indented
-Mandibular process of 1st branchial arch visible -3rd branchial arch visible |
13-20 |
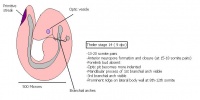
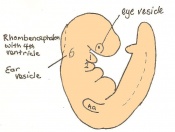
Following stage 14, The embryo continues to develop more distinguished features of a mouse, including forelimb and hindlimb bud, tail elongation, lungs and brain development. The stages 15-20 are summarised in the table below.
| Theiler Stage | Embryonic Age (dpc) | Stage Characteristic | Cell Characteristic | Somite Number(pairs) | |
|---|---|---|---|---|---|
| 15 | 9.5 (range 9-10.25)
Human carnegie stage: 12 |
Forelimb bud (8-12th somite pair)
-Posterior neuropore |
Hind limb bud
-Telencephalic and diencephalic vesicle division -Lung development -Dorsal pancreatic bud |
21-29 | |
| 16 | 10 (range 9.5-10.75)
Human carnegie stage: 13-15 |
Caudal neuropore closes
-Hind limb and tail bud |
Concave 3rd and 4th branchial arches.
-Rathke's pouch -Nasal processes -Ventral pancreatic bud |
30-34 | |
| 17 | 10.5 (range 10-11.25)
Human carnegie stage: 13-15 |
Deep Lens Indentation | Physiological umbilical hernia
-1st branchial arch (maxillary and mandibular components) -Brain tube development -Tail elongates |
35-39 | |
| 18 | 11 (range 10.5-11.25)
Human carnegie stage: 13-15 |
Lens Vesicle closes | Cervical somites not visible
-Brain rapidly grows -Nasal pit |
40-44 | |
| 19 | 11.5 (range 11-12.25)
Human carnegie stage: 16 |
Lens vesicle separated
–Closed and detached from ectoderm |
Eyes and their peripheral margins
-Limb-girdle and arm -Anterior footplate. -Auditory hillocks |
45-47 | |
| 20 | 12 (range 11.5-13)
Human carnegie stage: 17 |
Fingers | Anterior footplate (develops angles)
-Posterior footplate -Pigmentation of retina -Tongue and brain vesicles |
48-51 |
To visualize the mouse development, so far, a movie showing the mouse embryo can be seen by following the link [20]( note the heart beat) The last few stages of develop (stages 21-26) shown in table 5 are the complete prenatal developmental stages. These stages involve fine development of the pinna over the external acoustic meatus, finger and toe,hair, eyes and whiskers.
| Theiler Stage | Embryonic Age (dpc) | Stage Characteristic | Cell Characteristic | Somite Number(pairs) | |
|---|---|---|---|---|---|
| 21 | 13 (range 12.5-14)
Human Carnegie Stage 18-19 |
Anterior footplate indented
-Pinna develops |
Digits, elbow and wrist
-5 rows of vibrissae visible -Hair follicle over eye and ear -Lens loses lumen |
52-55 | |
| 22 | 14 (range 13.5-15)
Human Carnegie stage 20-23 |
Individual fingers on anterior footplate
-Umbilical hernia visible |
Deep indentations between toes( not separated)
-Long bones of limbs present -Hair present in pectoral, trunk and pelvis -Pinna turned forwards |
56-60 | |
| 23 | Human fetal period | Toes separate | Hair follicles present in cephalic region
-Eyelids open -Pinna covering ½ of external auditory meatus |
>60 | |
| 24 | 16 | Reposition of umbilical hernia | Parallel fingers (2-5)
-Toe nail primordia -Eyelids fused -Complete coverage by pinna -Increase in peritoneal sac size |
>60 | |
| 25 | 17 | Skin thickened and wrinkled
-Umbilical hernia disappeared |
Subcutaneous veins less visible
-Fingers and toes parallel -Whiskers visible |
||
| 26 | 18 | Skin thickened
Long whiskers |
Pinna larger (auditory meatus lumen not visible) |
To view the mouse embryo in late stages of development click the link [[21]] Following the prenatal development are the two Theiler stages 27 and 28 which involve birth of the mouse and postnatal development.
Timeline of Development
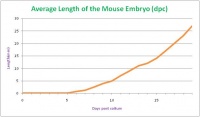
The development of the mouse embryo takes 19 days on average from fertilisation to newborn mouse. In comparison, the human embryo takes on average 40 weeks to progress from the one cell stage to newborn infant. This timing depends on the rate of development of the embryo. The average length of the mouse embryo varies during development depending on what stage the embryo is at and also on the degree of curvature to the body axis. The average length of the newborn embryo is between 23 and 27mm. The 26 Theiler stages are covered within 18 days, while stages 27 and 28 are covered during day 19 and the days after birth.
- Development of the Mouse Embryo
Genetics
The mouse is extensively used in experimental models because of its genome and its ability to be manipulated and studied.
Genome
In late 2002 the sequencing of the mouse genome was completed. Scientists have discovered that the haploid genome for the mouse is about 3 billion bases long having a total of 3000 Mb distributed over 20 chromosomes. The size of the mouse genome is closely related to the human genome size. Scientists estimate the gene count of the mouse genome to be 23,786 genes when compared to humans having 23,686 genes, a degree of size similarity can be shown.
The genetic map of the mouse
Genetic maps, the road maps of genetics, are of two types: linkage and physical. The 'sign posts' on the maps are loci, any location or marker in the genome that can be detected by genetic or DNA analysis. The term 'gene' is more restrictive than loci and refers to DNA segments that encode proteins or can be linked to phenotypes. Linkage maps are recombinational maps and are constructed by carrying out linkage crosses that measure the recombination frequency between genes or loci on the same chromosome.
- The first genetic linkage in the mouse (and first autosomal linkage in mammals) was described in 1915 in the classic paper on the linkage of pink-eyed dilution and albino.
- Genetic mapping with spontaneous mutations that created visible phenotypes, such as changes in coat colour/texture or behaviour was labarious and sometimes took years because crosses between mice carrying recessive mutations yielded so few informative progeny, and genes on only one or two chromosomes could be scored in each cross.
- The first real breakthrough in linkage mapping, enabling the scoring of many test markers and chromosomes in the same cross, was the discovery and use of co-dominant biochemical (isoenzyme) genes (e.g. glucose phosphate isomerase 1, Gpil; Hutton and Coleman 1969).
How large is the genome?
With the use of Feulgen reagent the quantitative DNA-specific staining can be achieved. Through micro photometric measurements of the staining intensity in individual sperm nuclei, it is possible to determine the total amount of DNA present in the haploid mouse genome. Measurements indicated a total haploid genome content of 3 pg, which translates into a molecular weight of 1.8 x 1012 daltons (Da).
How complex is the genome?
Another method for determining genome size relies upon the kinetics of DNA renaturation as a sign of the total content of different DNA sequences in a sample. When a solution of double stranded DNA is denatured into single strands which are then allowed to renature, the time required for renaturation is directly proportional to the complexity of the DNA in the solution, if all other parameters are held constant.
Complexity is a measure of the information contained within the DNA. The maximal information possible in a solution of genomic DNA purified from one animal or tissue culture line is equivalent to the total number of base pairs present in the haploid genome.The information content of a DNA solution is independent of the actual amount or concentration of DNA present. DNA obtained from one million cells of a single animal or cell line contains no more information than the DNA present in one cell. Furthermore, if sequences within the haploid genome are duplicates of one another — repeated sequences — the complexity will drop accordingly.
Renaturation analysis of mouse DNA reveals an overall complexity of approximately 1.3-1.8 x 109 bp. This value is only 40-60% of the size of the complete haploid genome and it implies the existence of a large fraction of repeated sequences.
What proportion of the genome is functional?
Bacterial species are remarkably efficient at packing the most genetic information into the smallest possible space. In one analysis of a completely sequenced 100 kb region of the E. coli chromosome, it was found that 84% of the total DNA content was actually used to encode polypeptides. Most of the remaining DNA is used for regulatory purposes, and only 2% was found to have no recognizable function.
Comparative sequence analysis over long regions of the mouse and human genomes shows evolutionary conservation over stretches of sequence that do not have coding potential or any obvious function. However, sequences can only be conserved when selective forces act to maintain their integrity for the benefit of the organism. Thus, conservation implies functionality.The fraction of the mouse genome that is functional is likely to lie somewhere between 5% and 10% of the total DNA present.
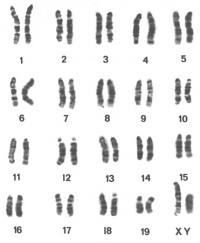
Chromosome number and banding patterns
All of the Mus musculus subspecies have the same standard karyotpye with;
- 20 pairs of chromosomes, including 19 autosomal pairs and the X and Y chromosomes
- All of the 19 autosomes as well as the X and Y chromosome appear to be telocentric, with a centromere at one end and a telomere at the other.
The biological explanation for this uniformity in chromosome morphology is entirely unknown; however, it makes the task of individual chromosome identification much more difficult than it is with human karyotypes.
Chromosome length and DNA content
The amount of DNA present in each chromosome can be estimated by measuring its length — cytologically — relative to the sum of the lengths of all 20 chromosomes and multiplying this fraction by the total genome length of 3,000 mb (Evans, 1989). From these measurements, one finds that the largest chromosome (1) has a DNA length of approximately 216 mb and the smallest chromosome (19) has a DNA length of 81 mb, with all others following in a near-continuum between these two values.
Mice with chromosomal aberrations
The diploid chromosomal complement of standard inbred laboratory strains is 2N=40:19 autosomes, X and Y chromosomes.The autosomes and the X chromosomes are telocentric (i.e. the centromere is at one end of a single-armed chromosome) while the y chromosome is acrocentric (i.e. it has a short p arm as well as the longer q arm, use of 'p' and 'q' is patterned on human chromosomal nomenclature). The sex determining genes resides in the short arm of the Y chromosome.
Strains of mice whose chromosomal complement devaites from the normal chromosomal makeup are designated chromosomal aberration starins. Chromosomal aberrations can include intra- and interchromosomal rearragements or aneuploidy. These include;
- inversions and transpositions, rearrangements of DNA segments within chromosomes
- reciprocal translocations, Robertsonian chromosomes, and insertions, exchanges of DNA segments between chromosomes
- aneuploidy, deviations from the normal diploid number of chromosomal arms in somatic cells (e.g. trisomies).
Some chromosomal deletions and duplications also may be cytologically detectable.
Comparative mapping
Comparative mapping began in the early 1970s and gained momentum until it culminated with the sequencing of the two genomes in 2001 and 2002.
- First conserved mouse and human autosomal linkage was reported in 1978.
- 13 conserved autosomal segments and estimated 178(+-)39 chromosomal rearrangements between mouse and human chromosomes were identified (Nadeau and Taylor,1984)
- Sequencing of the two genomes revealed that 95% of the coding sequence is conserved at the DNA level (Consortium,2002)
The Knockout Mouse
A knockout mouse is a genetically engineered mouse in which one or more genes have been turned off through a gene knockout. They are important animal models for studying the role of genes which have been sequenced. Mice are currently the most closely related laboratory animal species to humans for which the knockout technique can easily be applied.The first knockout mouse was created by Mario R, Capecchi, Martin Evans and Oliver Smilthies in 1989. [[22]]
- Use
Knocking out the acitivity of a gene provides information about what that gene normally does. Humans share many genes with mice. Consequently, observing the characterisitcs of knockout mice gives researchers information that can be used to better understand how a similar gene may contribute to disease in humans.
- Areas of research in which knockout mice have been useful include:
- cancer
- obesity
- heart disease
- diabetes
- arthritis
- anxiety
- aging
- Parkinson's disease.
- Limitations of the use of Knockout mice
While knockout mice technology represents a valuable research tool, its use may be limited. For example:
- About 15% of gene knockouts are developmentally lethal, which means that the genetically altered embryos cannot grow into adult mice. This problem is often overcome through the use of conditional mutations. The lack of adult mice limits studies to embryonic development and often makes it more difficult to determine a gene's function in relation to human health.
- Knocking out a gene may fail to produce an observable change in a mouse or may even produce different characteristics from those observed in humans in which the same gene is inactivated. E.g mutations in the p53 gene are associated with more than half of human cancers and often lead to tumors in a particular set of tissues. However, when the p53 gene is knocked out in mice, the animals develop tumors in a different array of tissues.
Examples of transgenic models for human diseases [[23]]
- Models for AIDS study
The HIV-1 virus is known to have two major receptors in human cells CD4 and CCR5. Transgenic rabbits expressing the human CD4 gene were used, the virus replicated in rabbit cells but was unable to generate any disease. Therefore, mouse models were generated. It appeared that transgenic rats expressing all the HIV-1 genes except gag and pol showed pathogeny having many similarities to human AIDS.
- Models for aging
Aging is a complex phenomenon which has only been partially described. Defects in genomes appear to be a major cause of aging. A growing number of trangenic models are being used to study aging. Mice in which the XPD gene has been knocked out are more sensitive to oxidative DNA damage. This sensitivity was increased further when the XPA gene was also knonked out. These models reflect some of the aging syndromes in human.
- Models for Cancer
Transgenic mice are used to generate models for cancer study. The first oncomouse expressed c-myc gene in the mammary gland. This was sufficient to trigger the formation of mammary tumors. Further studies made it pissible to identify additional genes involve in mammary cancer. Genes whose expression is amplified in mammary tumors have an oncogenic effect when used as transgenes. Crossing mice harboring different oncogenes and having knocked out genes has made it possible to determine the cooperative actions of some of these genes.
- The Future
The future of the mouse in genome analysis and as a model organism seems virtually unlimited. Whole genome sequence and gene preditction programs make it quite feasible to knock out or genetically modify every mouse gene. Almost certainly, ES cells will play a key role in future mouse genomics because they can be manipulated in culture. More phenotype screens that can detect mutations prior to making live mice will be needed to increase the number and types of mutations that can be detected in the ES cells themselves. While other model organisms, such as Drosophilia, yeast, worms and zebrafish may be easier to manipulate and allow analysis that require hundreds or thousands of animals, the mouse is likely to continue to be the premier mammalian model for understanding human inherited diseases.
International Mouse Strain Resource (IMSR)[24]
An on-line database that lists the international availability and supply of inbred, mutant, and genetically engineered mice.
Current Research
Overview
Mice are highly used models of developmental biology, immunity, neurobiology, and human diseases in pathology. It's main contribution to developmental biology has been through transgenic and knockout technology which are both very important to the research of early development or organogenesis (Slack, 2006).
2003: National Institute of Dental and Craniofacial Research (NIDCR)
Dr. Ashok Kulkarni and his colleagues [25] created a mouse model of the disease known in humans as dentinogenesis imperfecta III. This heredtitary tooth disorder involves the teeth wearing down to the innermost pulp.
They achieved this by knocking out the dentin sialophosphoprotein (Dspp) mouse gene, thought to be responsible for coordinating the mineralization of a tooth's dentin.
The role of Dspp is to organise the events during dentin mineralization, including potential regulation of proteoglycan levels. PMID for Dr. Ashok Kulkarni published article: 12721295
The International Knockout Mouse Consortium (IKMC)
There goal is to mutate all protein-coding mouse genes using a combination of gene trapping and gene targeting in mouse embryonic stem (ES) cells. Programs running under IKMC:
- Knockout Mouse Project (KOMP) (USA)
- European Conditional Mouse Mutagenesis Program (EUCOMM) (Europe)
- North American Conditional Mouse Mutagenesis Project (NorCOMM) (Canada)
- Texas A&M Institute for Genomic Medicine (TIGM) (USA)
2006: Green Lab, School of Medicine, Harvard University[26]
Researchers from the Green Lab cultivated Fibroblastic cells of a 12-13 day mouse embryo by 27 consecutive transfers and roughly 63 cell generations. This resulted in a new 'immortalised' cell line called, MMM which:
1.Supports the multiplication of the H9 cells better than the 3T3 line.
2.More effectively maintains the 'immortalised' cells as stem cells.
Link to published article: An immortalized drug-resistant cell line established from 12-13 day mouse embryos for the propagation of human embryonic stem cells
The Jackson Laboratory[27]
Researchers from The Jackson Laboratory conducted a series of transplantation experiments using three JAX® Mice strains: C57BL/6J (B6, 000664), B6.129S2-Alox5tm1Fun/J (004155), and B6.SJL-Ptprca Pepcb/BoyJ (002014) (also known as "pep boy").
They found that Alox5 deficiency blocks differentiation, alters the cell cycle, and induces apoptosis of long term LSCs (LT-LSCs). At the moment, it is not known why the Alox5 signaling pathway does not have the same effects on normal hematopoietic stem cells (HSCs).
Their results demonstrate that targeting a specific gene can completely inhibit only cancer stem cells in vivo.
The efficiency of an anti-stem cell strategy as a cancer therapy remains uncertain until clinical trials are undertaken in order to target ALOX5 in human leukemia patients (The Jackson Laboratory 2009).
2009: Department of Experimental Medicine, Histology and Embryology Unit, University of Pavia, Pavia, Italy
Cornaglia et al. used the knockout mouse to research how mutations in the diastrophic dysplasia sulphate transporter (dtdst) gene caused different forms of the disease chondrodysplasia in humans. By producing a knockout mice strain of this mutated dtdst gene, they were able to investigate how this gene affected tissue organisation, matrix structure and cellular differentiation in the epiphyseal growth plate of the bone.
Link to published article: Dysplastic histogenesis of cartilage growth plate by alteration of sulphation pathway: a transgenic model.
A Pubmed search of the use of the mouse as a model in the study of embryology was done which produced 5213 literature articles. To view this search please click here . A brief selection of these articles include:
2009: Gabriela Pavlinkova, J Michael Salbaum and Claudia Kappen
They investigated how maternal diabetes alters transcriptional programs in the developing embryo by using a mouse diabetes model. Maternal diabetes is a known risk factor for abnormalities in the newborn, but it is unknown how this comes about.
They hypothesized that the defects were caused by a change in developmental pathways that can result in changes in gene expression. After experimental results were obtained, it was found that exposure to maternal diabetes had caused alterations in the transcriptional profiles of the developing embryo.
Link to published artile: Maternal diabetes alters transcriptional programs in the developing embryo.
2009: Division of Pediatric Otolaryngology - Head and Neck Surgery at the Cincinnati Children's Hospital, Cincinnati, Ohio
Ravindhra et al. hypothesized that the rate and direction of chondrocyte growth was affected by Fibroblast growth factor 18 (FGF18). To prove this, they used the mouse as a model and observed the affect that FGF18 had on the cartilage specifying gene, Sox9. There results found that GF18 did regulate the rate and direction of chondrocyte growth by up-regulating Sox9 expression.
Link to published article: Fibroblast growth factor 18 gives growth and directional cues to airway cartilage.
2009:Department of Veterinary Biosciences, College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Illinois & the Department of Cell Biology, School of Medicine, University of Virginia, Charlottesville, Virginia
Denise, et al. found that the epithelial-mesenchymal interactions in the development of the Wolffian duct and testis cord are critical for normal development. They were able to identify Inhba and its protein product activin A as being an interstitially derived factor that acts upon sertoli cells in the testis that hints that crosstalk occurs between both organs.
Link to published article: Epithelial-mesenchymal crosstalk in Wolffian duct and fetal testis cord development.
2009: Laboratorio di Biologia Molecolare, Istituto G Gaslini [28], Genova, Italy
Fardin et al. used the mouse as a model for understanding the function of oncogenes involved in epithelial-mesenchymal uncontrolled growth (Fardin et.al. 2009).
They examined a protein Dbl which belongs to the family of proto-oncogenes or proteins involved in normal cell growth. These proteins, when mutated, become oncogenes and cause unregulated cell growth. The transgenic mice were used to incorporate the complement DNA strand of Dbl oncogene for analysis.
There results showed that the expression of onco-Dbl in mice correlated with up regulation of epithelial transition, apoptosis, vasculogenesis and cell proliferation.
2009: Department of Microbiology and Immunology, University of Maryland School of Medicine[29], Baltimore, MD, USA
Andreasen et al. used the mouse as a model of immune response to respiratory tract infections by the bacteria Bordetella pertussis.
They infected the mouse with the bacteria, and found that the mouse secretes a virulence factor called pertussis toxin. This toxin is responsible for promoting bacterial growth and proliferation in the airways.
The study showed that the toxin ,rather than the bacterial cell numbers, is important in the production of cytokine and chemokine responses, and hence the pertussis toxin produces the host immune response not the bacteria.'
2009: Regales, et al.
They used transgenic mice as models for testing anticancer drugs in lung tumours involved with tyrosine receptor mutations. The lung tumours' mutations were of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance mutations T790M. By doing this, they were able to evaluate the effectiveness of a variety of anticancer drugs.
In addition, they tested two anticancer agents, BIBW-2992 tyrosine kinase inhibitor and EGFR specific antibody cetuximab, on the mice containing EGFR mutated lung tumors. It was found that the combination of the two agents produced marked shrinkage of tumours with T790M mutations.
Link to published article: Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer.
2009: Department of Virology, Statens Serum Institut, Copenhagen, Denmark
Gram et al. used mice in the development of HIV-1 vaccines. Note that a vaccine needs to have several factors including several antigens and epitopes, as well as an appropriate adjuvant capable for inducing a strong host cellular immune response.
They used transgenic mice to develop a new lipophillic adjuvant called CAF01, which has been studied to assist in generating antibody and CD4 T helper cell responses. This process was found to generate CD8 T lymphocytes against HIV-1 derived cytotoxic T lymphocyte epitope peptides in human leukocyte antigen mouse models.
Link to published article:A novel liposome-based adjuvant CAF01 for induction of CD8(+) cytotoxic T-lymphocytes (CTL) to HIV-1 minimal CTL peptides in HLA-A*0201 transgenic mice.
Glossary
Allantoic bud: A sac or diverticulum that prodrudes out from the caudal wall of the umbilical vesicle into the connecting stalk.
Allophenic: A word used to refer to an animal consisting with a minimum of two genetically-different population of cells originating from different embryos.
Amnion: A fluid filled sac which encloses the amniotic cavity.
Ampulla: The dilated end of the Fallopian tube, site of fertilization.
Auricular hillocks: Embryological bumps which correspond to external ear features in adulthood
Blastocyst: Structure formed when morula develops a fluid filled space( blastocystic cavity). The balstocyst contains trophoblast and embryoblast cells.
Branchial arch: Primordial pharyngeal structures, also known as pharyngeal arch
Cardiogenic plate: Consisting of cardiogenic mesoderm where the heart primordium will develop.
Cephalic: Term used to describe the head.
Chimera: An animal consisting with a minimum of two genetically-different population of cells originating from different zygotes.
Corpus Luteum: The yellowish structure formed as a result of the ruptured graafian follicle that contains cells secreting Progesterone.
Diencephalon: The third ventricle consisting of 3 swellings which contains the embryonic neural tube region that will form the hypothalmus, thalmus and epithalamus of the brain.
Ectoderm:One of the 3 germ cell layers, which gives rise to the epidermis, central and peripheral nervous system, ear, eye and neural crest cells.
Ectoplacental cone: Term describing the polar trophectoderm cell proliferation, of the bastocyst.
Endoderm:One of the 3 germ cell layers which gives rise to epithelial lining of the respiratory and GIT tract, accessory organs of GIT and lines the yolk sac.
Epiblast: A thick layer of columnar cells related to the amniotic cavity.
Estrous cycle: The cycle of female animals which involves the alterations in the female tract as well as sexual receptivity in relation to hormone changes.
Exocoelom: Term describing the yolk sac, the source of nutrients and cells for the developing embryo.
External auditory meatus: Also know as ear canal, is the tube from the outer ear to the inner ear.
Footplate: The paddle shaped structure which forms in the anterior and posterior limb buds to form the hand and foot.
Forelimb: The anterior limb of an animal.
Gastrulation: The process in which the three germ layers are developed. These germ layers are the precursors of all embryoni tissue and axial orientation.
Genome: The total genetic content or genes in an animal.
Gestation: The period of development from when fertilization occurs or conception until time of birth.
Haploid: A set of chromosomes containing only one member of each chromosome pair. The sperm and egg are haploid.
Heart rudiment: Cone like transitional structure of migrating myocardial precursors which form the primitive heart tube.
Hindlimb: The posterior limb of an animal.
Homology: Anatomical structure or traits of two different organisms which resemble similarity from a common ancestral organism.
Mesoderm: One of the 3 germ cell layers, which gives rise to all skeletal muscles, blood cells and vessel lining, linings of all body cavities, ducts and organs of reporductive system and most of cardiovascular system.
Morulla: Embryonic structure which consists of 12 to 32 blastomeres.
Mutation: is a randomly derived change to the nucleotide sequence of the genetic material of an organism.
Neural plate:Structure when embryonic ectoderm thickens and forms an elongated plate of thickened epithelial cells.
Neural tube: Structure when neural folds move together and fuse, converting the neural plate into a tube.
Neuropore: The temporary opening of the neural tube at both rostral and caudal ends of the embryo.
Oncogenic: An oncogene is a gene that, when mutated or expressed at high levels, helps turn a normal cell into a cancer cell.
Optic pit: Depression for optic disc.
Optic placode: The surface structure of ectoderm thickening in the head region of an early embryo, forms a component of the sensory system of the eye.
Ovulation: The process of releasing an egg or oocyte from the ovary ( normally midcycle).
Pancreatic bud: Structure which forms the pancreas between layers of mesentery (dorsal and ventral).
Peritoneal sac:Space like structure of the peritoneal cavity. Can be divided into greater and lesser sacs.
Pinna: The external part of the ear. Functions to amplify sound by directing sound waves into the ear.
Polyestrous: Having many estrous cycles during a breeding season.
Postnatal:The period of time after birth of a fetus.
Prenatal:The period of time before birth of a fetus
Pre-somites:The structure before the appearance of somite pairs.
Reicherts membrane:A thick basement membrane structure in the parietal wall of the yolk sac.
Renaturation: The process by which proteins or complementary strands of nucleic acids reform their native conformations.
Somatic tissue: Tissue that is related to the body of an animal.
Somite: Mesoderm structure formed from paraxial mesoderm which differentiates to form the sclerotome and dermamyotome. The somites form the muscle and connective tissue.
Stem Cell: An undifferentiated cell that has the potential to become differentiated. They can retain the ability to divide throughout life.
Subcutaneous:The soft tissue structure underlying the epidermis or skin.
Telencephalon: The structure of the embryonic neural tube region which forms the cerebral hemispheres, formed from division of the forebrain.
Telocentric: a chromosome like a straight rod with the centromere in terminal position.
Transgenic:A genetically modified organism (GMO) or genetically engineered organism (GEO) is an organism whose genetic material has been altered using genetic engineering techniques.
Transposition: is a kind of mutation in which a chromosomal segment is transfered to a new position on the same or another chromosome.
Trophectoderm:The cell layer which differentiates to form the trophoblast.
Trophoblast: Outer cell layer of blastocyst which gives rise to embryonic part of the placenta.
Umbilical hernia:The outward protrusion of the abdominal lining or part of the abdominal organ at the umbilicus.
Vibrissae:Follicles of hair also called whiskers.
Zona pellucida:A specialised zone of cells surrounding the oocyte and blastocyst during early development. Functions in allowing sperm to bind and prevents polyspermy.
(Moore & Persaud, 2008; Biology Online, accessed 2009 [30])
References
Books and Articles
- Andreasen, C., Powell, D.A. & Carbonetti, N.H. (2009). Pertussis Toxin Stimulates IL-17 Production in Response to Bordetella pertussis Infection in Mice. PLoS ONE, 4(9), 1-11. Accessed from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2738961.
- Andrews, E.A. (1895). Embryology. The American Naturalist, 29, 167-169.
- Archambeault, D. R., Tomaszewski, J., Joseph, A., Hinton, B. T., & Yao, H. (2009). Epithelial-mesenchymal crosstalk in Wolffian duct and fetal testis cord development. Genesis, 47(1), 40-48.
- Bard, J.B.L., Kaufman, M.H., Dubreuil, C., Brune, R.M., Burger, A., Baldock, R.A., & Davidson, D.R. (1998). An internet accessible database of mouse development anatomy based on a systemic nomenclature. Mechanisms of Development, 74, 111-120.
- Bhamrah, H., & Juneja, K. (2003). Molecular Cell Biology 1st edition. New Delhi. Anmol Publications pp.1-416.
- Cornaglia, A., Casasco, A., Casasco, M., Riva, F., & Necchi, V. (2009). Dysplastic histogenesis of cartilage growth by alteration of sulphation pathway: a transgenic model. Connective Tissue Research, 50(4), 232-42.
- Elluru, R G., Thompson, F., & Reece, A. (2009). Fibroblast Growth Factor 18 Gives Growth and Directional Cues to Airway Cartilage. The Laryngoscope Journal, 199(6), 1154 - 1165. PMID: 19358209.
- Fardin, P., Ognibene, M., & Vanni, C. (2009). Induction of Epithelial Mesenchimal Transition and Vasculogenesis in the Lenses of Dbl Oncogene Transgenic Mice. PLoS ONE, 4(9), 1-15. Accessed from, http://assets0.pubget.com/pdf/19759912.pdf
- Ginsburg, M., Snow, M.H., & McLaren, A. (1990). Primordial germ cells in the mouse embryo during gastrulation. Development, 110, 521-528.
- Gram, G.J., Karlsson, I., Agger, E.M., Andersen, P., & Fomsgaard, A. (2009). A Novel Liposome-Based Adjuvant CAF01 for Induction of CD8+ Cytotoxic TLymphocytes(CTL) to HIV-1 Minimal CTL Peptides in HLA-A 0201 Transgenic Mice. PLoS ONE 4(9), 1-5. Accessed from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2736401
- Greenwald, G. (1956). The Reproductive Cycle of the Field Mouse, Microtus californicus. Journal of Mammalogy, 37(2), 213-222.
- Hedrich, H., Bullock, G., & Petrusz, P. (2004). The Laboratory Mouse. London. Elsevier Academic Press pp.3-24. Accessed from, http://www.google.com/books?id=XLIarRWHikAC&printsec=frontcover&dq=The+laboratory+mouse#v=onepage&q=&f=false
- Hill, M. (2009). The Mouse, UNSW Embryology. ISBN: 978 0 7334 2609 4. Accessed from https://embryology.med.unsw.edu.au/embryology/index.php/Mouse_Development
- Moore, K., & Persaud, T. (2008). The Developing Human Clinically Oriented Embryology 8th Edition. Philadelphia. Saunders Elsevier.
- Nagy, A., Gertsenstein, M., Vintersten, K., & Behringer, R. (2003). Manipulating the Mouse Embryo: A Laboratory Manual 3rd edition. New York. Cold Spring Harbor Laboratory Press pp.1-100.
- Pavlinkova, G., Salbaum, J.M. & Kappen, C. (2009). Maternal diabetes alters transcriptional programs in the developing embryo. BMC Genomics, 10(274), 1-12. Accessed from http://www.biomedcentral.com/content/pdf/1471-2164-10-274.pdf
- Regales, Gong, Y., Shen, R., Stanchina, E., Vivanco, I., Goel1, A., Koutcher, J., Spassova, M., Ouerfelli, O., Mellinghoff, I., Zakowski, M., Politi, K., & Pao, W. (2009). Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutation in lung cancer. Journal of Clinical Investigation. Accessed from http://www.jci.org/articles/view/38746
- Rouvroit, C. L., & Goffinet, A. M. (1998). Advances in Anatomy, Embryology, and Cell Biology: The Reeler Mouse as a Model of Brain Development. Berlin. Springer.
- Shiro, I., Meytha, M., Cristina, V., Karen, E., Walid, K., Howard, G. (2006). An immortalized drug-resistant cell line established from 12-13-day mouse embryos for the propagation of human embryonic stem cells. Differentiation. Accessed from http://pt.wkhealth.com/pt/re/diff/abstract.00003444-200604000-00003.htm;jsessionid=KJLDc4JX2vKFKSQbLw7SSJTx2pGy4dMvGQJ9zpGxDdZ5hN1YyC6H!-2135654213!181195629!8091!-1
- Silver, L. M. (1995). Mouse Genetics: Concepts and Applications. Bar Harbor. Oxford University Press. Accessed from http://www.informatics.jax.org/silver/
- Theiler, K. (1989). The House Mouse: Atlas of Embryonic Development. New York. Springer - Verlag. Accessed from http://genex.hgu.mrc.ac.uk/Databases/Anatomy/new/
Websites
- Bard, J., & Kaufman, M. (2003). The Edinburgh Mouse Atlas Project. The Medical Research Council & University of Edinburgh, Accessed from http://genex.hgu.mrc.ac.uk/Atlas/intro.html
- Eye on DNA. Image of Transgenic Mouse Embryo Wins Nikon Small World Photomicrography Competition, retrieved September 24, 2009. http://www.eyeondna.com/2007/10/08/image-of-transgenic-mouse-embryo-wins-nikon-small-world-photomicrography-competition/
- Geneall. Dame Anne Laura Dorinthea McLaren, retrieved September 21, 2009. http://www.geneall.net/U/per_page.php?id=557208
- Harvard University. Howard Green, M.D., retrieved September 24, 2009. http://cellbio.med.harvard.edu/faculty/green/index.html
- Institute of Medical Biology. Davor Solter, retrieved October 12, 2009. http://www.imb.a-star.edu.sg/Default.aspx?tabid=224
- International Knockout Mouse Consortium. About the International Knockout Mouse Consortium, retrieved September 19, 2009.
- MedicineNet. MedTerms Dictionary, retrieved October 10, 2009. http://www.medterms.com/script/main/hp.asp
- Memorial University. Oration honouring Wesley Whitten, retrieved September 11, 2009. http://www.mun.ca/marcomm/gazette/2000-2001/june14/convocation8.html
- Merriam-Webster Online Medical Dictionary. Retrieved September 23, 2009. http://www.merriam-webster.com/medical/allophenic
- National Institute of Health Record. NIDCR Team Creates Mouse Model of Disease, retrieved September 19, 2009. http://nihrecord.od.nih.gov/newsletters/09_16_2003/scinews.htm
- The BioTech Weblog. Mouse Resequencing and SNP Discovery Project Completed, retrieved September 24, 2009. http://www.biotech-weblog.com/50226711/mouse_resequencing_and_snp_discovery_project_completed.php
- The Jackson Laboratory. Novel Leukemia Treatment Discovered in Mice, retrieved September 21, 2009. http://jaxmice.jax.org/news/2009/leukemia.html
- University of Iowa : College of Liberal Arts and Sciences. Alumni Fellows, retrieved September 26, 2009. http://images.google.com.au/imgres?imgurl=http://www.clas.uiowa.edu/_includes/images/alumni/beatrice.jpg&imgrefurl=http://www.clas.uiowa.edu/alumni/programs/fellows/2002/&usg=__ZTVwhRDWmWlpQWiTgm7QPANk0ow=&h=218&w=182&sz=10&hl=en&start=2&sig2=WxT11tEDMuSTsbeT1PzXjQ&um=1&tbnid=7WxPUu34U_wdOM:&tbnh=107&tbnw=89&prev=/images%3Fq%3Dbeatrice%2Bmintz%26hl%3Den%26sa%3DN%26um%3D1&ei=ECy2Sp_JKMiGkAX9k9XBCw
- Wikipedia. Chimera (genetics), retrieved October 10, 2009. http://en.wikipedia.org/wiki/Chimera_(genetics)
- Wikipedia. Gregor Mendel, retrieved September 20, 2009. http://en.wikipedia.org/wiki/File:Gregor_Mendel.png
- Wikipedia. Rudolf Jaenisch, retrieved September 17, 2009. http://en.wikipedia.org/wiki/File:Jaenisch_2003_by_Sam_Ogden.jpg
ANAT2341 group projects
Project 1 - Rabbit | Project 2 - Fly | Project 3 - Zebrafish | Group Project 4 - Mouse | Project 5 - Frog | Students Page | Animal Development