2009 Group Project 4
THE MOUSE
Mus musculus
Introduction
The mouse is a small animal which can be effectively used as a model to study embryological development. It belongs to the class mammalian and order rodentia and shares a significant similarity of homology with humans. The mouse is one of the most commonly used animals as model organisms in experimental embryology and most sciences. The mouse is a useful mammalian model for human embryological development because the mouse;
- Has the same size genome as the human genome
- Genes can be easily manipulated and studied
- Have a high degree of homogeny with humans
- Produces a large offspring in a short amount of time
- Are not limited in ethical issues as they can be manipulated in ways which would be unethical to do to humans
- Are small organisms and are easily maintained
- Are not expensive
Timeline of Development
Edinburgh Atlas Project[1]
- Development of the Mouse Embryo
Staging
Mouse embryonic development commences once the female's egg or oocyte has become fertilized by the male's sperm. Mouse development has a gestation period of 19-21 days, and can range in different strains of mice. The development of an embryo can be categorized into different stages including cell number, somite stages and morphology. Link to a movie showing mouse embryo [2] The most common method of staging is by Theiler (1989) which categorizes mouse development into prenatal and postnatal stages consisting of 26 and 2 stages respectively. Downs and Davies (1993) have also established a method of staging the mouse development based on morphological changes. Link to Downs and Davies[3]
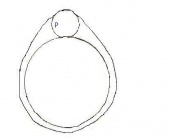
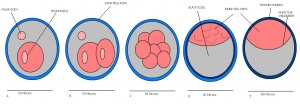
The developmental stages of the mouse embryo can be summarised in the tables and corresponding figures below in relation to characteristics at that stage. The first stage begins with fertiliation of the egg which divides into multiple cells to form a morulla and further go on to form a blastocyst which is ready for implantation.
| Theiler Stage | Embryonic Age in Days Post Coitum (dpc) | Stage Characteristic | Cell Characteristics | Zona Pellucida | Location | |
|---|---|---|---|---|---|---|
| 1 | 0-0.9
(range 0-0.25) |
One-celled embryo (fertilized) | One cell | Present | Ampulla | |
| 2 | 1
(range 1-2.5) |
Dividing egg | 2-4 cells
- 1st cleavage after 24hrs |
Present | Travelling down oviduct | |
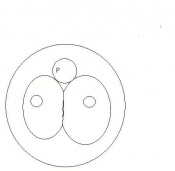
| 3 | 2
(range 1-3.5) |
Morula (early to fully compacted) | 4-16 cells | Present | Oviduct (utero-tubal junction) | |
| 4 | 3
(range 2-4) |
Morula to Blastocyst
-Intra-cellular matrix -Blastocoelic cavity |
16-40 compacted cells
-Inner cell mass -Outer layer of trophectoderm cells |
Present | Uterine lumen | |
| 5 | 4
(range 3-5.5) |
Zona free Blastocyst (hatching) | Blastocyst implants as zona pelludica is lost | Absent | Uterine lumen |
Dr Mark Hill 2009, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G
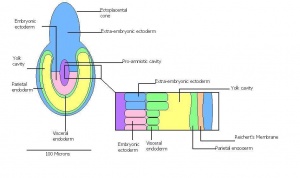
Once the Blastocyst has lost the zona pellucida, it is free to implant onto the uterine wall. After implantation multiple features of a trilaminar embryo start to develop. Ectoderm, endoderm and mesoderm layers form, which further develop many structures corresponding to different stages as summariesed in the table below. The developing mouse embryo which is implanted on the uterine wall can be viewed by ultrasonic radiation. Link to ultrasound of mouse embryo [4]
| Theiler Stage | Embryonic age in Days Post Coitum (dpc) | Stage Characteristic | Cell characteristics | |
|---|---|---|---|---|
| 6 | 4.5 (range 4-5.5)
Human carnegie stage: 4 |
Attachment of blastocyst
-Implantation |
Embryonic Endoderm | |
| 7 | 5 (range 4.5-6)
Human carnegie stage: 5 |
Implantation
-Egg cylinder -Ectoplacental cone |
Inner cell mass increases
-Epiblast -Mural trophectoderm lined by endoderm | |
| 8 | 6 (range 5-6.5)
Human carnegie stage: 5 |
embryonic and extra-embryonic regions
-Pro-amniotic cavity |
Trophoblast giant cells invade
-Maternal blood invades -Reichert's membrane | |
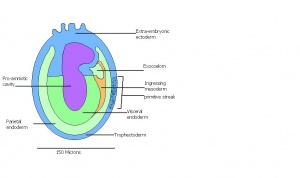
| 9 a) | Pre-streak | Advanced Endometrial and egg cylinder
-Embryonic axis |
Embryonic and extra-embryonic ectoderm
-Uterine crypts lose lumen | |
| 9 b) | Early streak | Gastrulation begins | Mesodermal cells | |
| 10 a) | 7 (range 6.5-7.5)
Mid to late streak Human carnegie stage: 8 |
Amnion formation | Amniotic fold
-Allantoic bud -Primitive node -Amnion closes | |
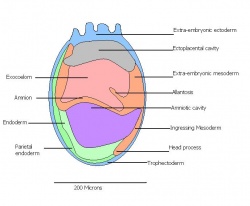
| 11 | 7.5 (range 7.25-8)
Human carnegie stage: 9 |
Neural plate and presomites | Amniotic cavity, exocoelom and ectoplacental cleft
-Allantoic bud elongates -Notochodal plate -Early head fold -Foregut pocket |
This table can be seen in more detail by following the link More on stage 6-11
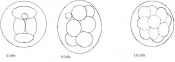
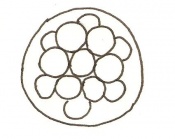
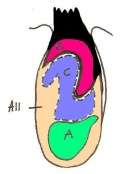
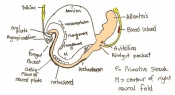
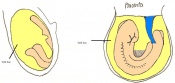
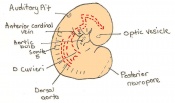
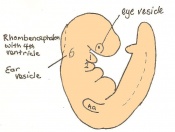
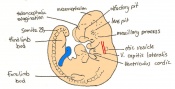
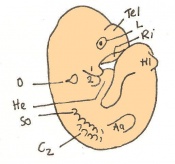
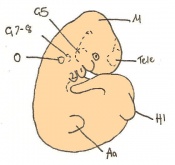
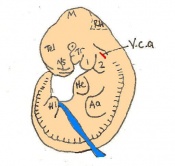
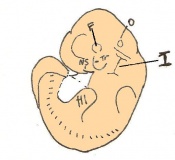
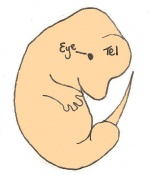
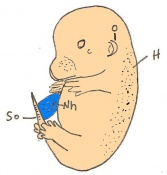
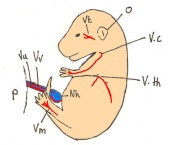
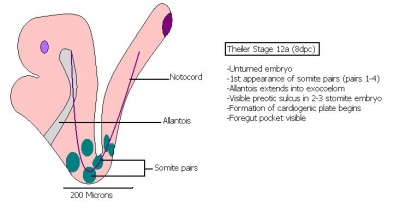
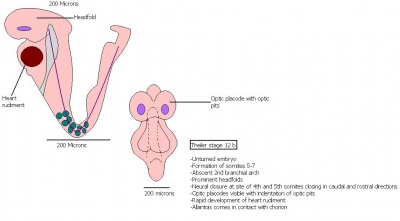
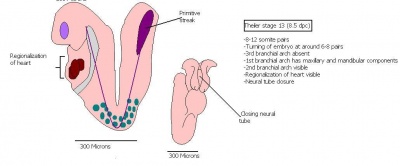
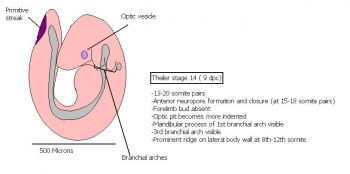
The next stage (Theiler stage 12) is marked by the first somite pair formation. Stages 13 onwards is characterized by increasing somite pairs up to 20 somite pairs. These embryo stages have characteristic features of developing forgut, prominent head fold, optic pits, branchial arch and neuropore formation. These four stages can be summarised in the figures 4-7 below. These stages can be seen in table form by following the link Table of stages 12-14
Table 3: Mouse embryonic stages from Theiler stage 15 to 20 ( somite stages)
| Theiler Stage | Embryonic Age (dpc) | Stage Characteristic | Cell Characteristic | Somite Number(pairs | |
|---|---|---|---|---|---|
| 15 | 9.5 (range 9-10.25)
Human carnegie stage: 12 |
Formation of Forelimb bud
-Posterior neuropore |
8-12th somite pair condensation of forelimb bud is visible
-Hind limb bud appears -Forebrain vesicle division into telencephalic and diencephalic vesicles -Lung development commences -1st sign of Pancreas morphogenesis of dorsal pancreatic bud (22-25 somites). |
21-29 | |
| 16 | 10 (range 9.5-10.75)
Human carnegie stage: 13-15 |
Caudal neuropore closes
-Hind limb bud (23rd-28th somite) and tail bud |
Concave 3rd and 4th branchial arches.
-Rathke's pouch formation -Nasal processes formation. -Ventral pancreatic bud appears |
30-34 | |
| 17 | 10.5 (range 10-11.25)
Human carnegie stage: 13-15 |
Deep Lens Indentation | Lens pit is deepened and has a narrowed outer opening.
-Physiological umbilical hernia present. -1st branchial arch divides into maxillary and mandibular components. -Advanced development of brain tube -Tail elongates and thins |
35-39 | |
| 18 | 11 (range 10.5-11.25)
Human carnegie stage: 13-15 |
Closure of Lens Vesicle | Cervical somites no longer visible
-Brain rapidly grows -Formation of nasal pit |
40-44 | |
| 19 | 11.5 (range 11-12.25)
Human carnegie stage: 16 |
Lens vesicle separated completely from surface
–Closed and detached from ectoderm |
Well Defined eyes and their peripheral margins
-Forelimbs divided into two regions -Proximal part of the future limb-girdle and 'arm' -Peripheral part forming a circular or anterior footplate. -Otic pit medial and lateral margins move together -Auditory hillocks visible |
45-47 | |
| 20 | 12 (range 11.5-13)
Human carnegie stage: 17 |
First sign of fingers | Anterior footplate no longer circular (develops angles)
-Posterior footplate visible -Pigmentation of retina visible -Tongue and brain vesicles identifiable |
48-51 |
History of the use of the Mouse Embryo Model
Genetics
Genome
Sequencing of the mouse genome was completed in late 2002. Tha haploid genome is about 3 billion long (3000 Mb distributed over 20 chromosomes) and therefore equal to the size of the human genome. The current estimated gene count is 23,786 and humans are estimated to have 23,686 genes.
The genetic map of the mouse
Genetic maps, the road maps of genetics, are of two types: linkage and physical. The 'sign posts' on the maps are loci, any location or marker in the genome that can be detected by genetic or DNA analysis. The term 'gene' is more restrictive than loci and refers to DNA segments that encode proteins or can be linked to phenotypes. Linkage maps are recombinational maps and are constructed by carrying out linkage crosses that measure the recombination frequency between genes or loci on the same chromosome.
- The first genetic linkage in the mouse (and first autosomal linkage in mammals) was described in 1915 in the classic paper on the linkage of pink-eyed dilution and albino (Haldane et al., 1915)
- Genetic mapping with spontaneous mutations that created visible phenotypes, such as changes in coat colour/texture or behaviour was labarious and sometimes took years because crosses between mice carrying recessive mutations yielded so few informative progeny, and genes on only one or two chromosomes could be scored in each cross.
- The first real breakthrough in linkage mapping, enabling the scoring of many test markers and chromosomes in the same cross, was the discovery and use of co-dominant biochemical (isoenzyme) genes (e.g. glucose phosphate isomerase 1, Gpil; Hutton and Coleman 1969).
How large is the genome?
With the use of Feulgen reagent the quantitative DNA-specific staining can be achieved. Through micro photometric measurements of the staining intensity in individual sperm nuclei, it is possible to determine the total amount of DNA present in the haploid mouse genome (Laird, 1971). Measurements indicated a total haploid genome content of 3 pg, which translates into a molecular weight of 1.8 x 1012 daltons (Da).
How complex is the genome?
Another method for determining genome size relies upon the kinetics of DNA renaturation as a sign of the total content of different DNA sequences in a sample. When a solution of double stranded DNA is denatured into single strands which are then allowed to renature, the time required for renaturation is directly proportional to the complexity of the DNA in the solution, if all other parameters are held constant.
Complexity is a measure of the information contained within the DNA. The maximal information possible in a solution of genomic DNA purified from one animal or tissue culture line is equivalent to the total number of base pairs present in the haploid genome.The information content of a DNA solution is independent of the actual amount or concentration of DNA present. DNA obtained from one million cells of a single animal or cell line contains no more information than the DNA present in one cell. Furthermore, if sequences within the haploid genome are duplicates of one another — repeated sequences — the complexity will drop accordingly.
Renaturation analysis of mouse DNA reveals an overall complexity of approximately 1.3-1.8 x 109 bp. This value is only 40-60% of the size of the complete haploid genome and it implies the existence of a large fraction of repeated sequences.
Comparative mapping
Comparative mapping began in the early 1970s and gained momentum until it culminated with the sequencing of the two genomes in 2001 and 2002.
- First conserved mouse and human autosomal linkage was reported in 1978.
- 13 conserved autosomal segments and estimated 178(+-)39 chromosomal rearrangements between mouse and human chromosomes were identified (Nadeau and Taylor,1984)
- Sequencing of the two genomes revealed that 95% of the coding sequence is conserved at the DNA level (Consortium,2002)
Current Research
References
1. Dr Mark Hill 2009, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G [5]
2. Bard, Kaufman, Dubreuil, Brune, Burger, Baldock, Davidson (1998). An internet accessible database of mouse development anatomy based on a systemic nomenclature, Mechanisms of development, (74) 111-120.
3. Theiler, K 1989. 'The House Mouse'. Springer- Verlag, New York
note: reference edinburgh atlas project
ANAT2341 group projects
Project 1 - Rabbit | Project 2 - Fly | Project 3 - Zebrafish | Group Project 4 - Mouse | Project 5 - Frog | Students Page | Animal Development