Mouse Knockout: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| Line 23: | Line 23: | ||
|-bgcolor="F5FAFF" | |-bgcolor="F5FAFF" | ||
| | | | ||
* '''Restoration of Spermatogenesis and Male Fertility Using an Androgen Receptor Transgene'''<ref name="PMID25803277"><pubmed>25803277</pubmed></ref> "Androgens signal through the androgen receptor (AR) to regulate male secondary sexual characteristics, reproductive tract development, prostate function, sperm production, bone and muscle mass as well as body hair growth among other functions. We developed a transgenic mouse model in which endogenous AR expression was replaced by a functionally modified AR transgene. A bacterial artificial chromosome (BAC) was constructed containing all AR exons and introns plus 40 kb each of 5' and 3' regulatory sequence. Insertion of an internal ribosome entry site and the EGFP gene 3' to AR allowed co-expression of AR and EGFP. Pronuclear injection of the BAC resulted in six founder mice that displayed EGFP production in appropriate AR expressing tissues. The six founder mice were mated into a Sertoli cell specific AR knockout (SCARKO) background in which spermatogenesis is blocked at the meiosis stage of germ cell development. The AR-EGFP transgene was expressed in a cyclical manner similar to that of endogenous AR in Sertoli cells and fertility was restored as offspring were produced in the absence of Sertoli cell AR. Thus, the AR-EGFP transgene under the control of AR regulatory elements is capable of rescuing AR function in a cell selective, AR-null background. These initial studies provide proof of principle that a strategy employing the AR-EGFP transgene can be used to understand AR functions." | |||
* '''Mouse library set to be knockout'''<ref><pubmed>21677718</pubmed>| [http://www.nature.com/news/2011/110615/full/474262a.html Nature News]</ref> "Launched in 2006 in North America and Europe, the effort aims to disable each of the 20,000-odd genes in the mouse genome and make the resulting cell lines available to the scientific community." | * '''Mouse library set to be knockout'''<ref><pubmed>21677718</pubmed>| [http://www.nature.com/news/2011/110615/full/474262a.html Nature News]</ref> "Launched in 2006 in North America and Europe, the effort aims to disable each of the 20,000-odd genes in the mouse genome and make the resulting cell lines available to the scientific community." | ||
* '''A conditional knockout resource for the genome-wide study of mouse gene function'''<ref name="PMID21677750"><pubmed>21677750</pubmed></ref> "Gene targeting in embryonic stem cells has become the principal technology for manipulation of the mouse genome, offering unrivalled accuracy in allele design and access to conditional mutagenesis. To bring these advantages to the wider research community, large-scale mouse knockout programmes are producing a permanent resource of targeted mutations in all protein-coding genes. Here we report the establishment of a high-throughput gene-targeting pipeline for the generation of reporter-tagged, conditional alleles. Computational allele design, 96-well modular vector construction and high-efficiency gene-targeting strategies have been combined to mutate genes on an unprecedented scale. So far, more than 12,000 vectors and 9,000 conditional targeted alleles have been produced in highly germline-competent C57BL/6N embryonic stem cells. High-throughput genome engineering highlighted by this study is broadly applicable to rat and human stem cells and provides a foundation for future genome-wide efforts aimed at deciphering the function of all genes encoded by the mammalian genome." | * '''A conditional knockout resource for the genome-wide study of mouse gene function'''<ref name="PMID21677750"><pubmed>21677750</pubmed></ref> "Gene targeting in embryonic stem cells has become the principal technology for manipulation of the mouse genome, offering unrivalled accuracy in allele design and access to conditional mutagenesis. To bring these advantages to the wider research community, large-scale mouse knockout programmes are producing a permanent resource of targeted mutations in all protein-coding genes. Here we report the establishment of a high-throughput gene-targeting pipeline for the generation of reporter-tagged, conditional alleles. Computational allele design, 96-well modular vector construction and high-efficiency gene-targeting strategies have been combined to mutate genes on an unprecedented scale. So far, more than 12,000 vectors and 9,000 conditional targeted alleles have been produced in highly germline-competent C57BL/6N embryonic stem cells. High-throughput genome engineering highlighted by this study is broadly applicable to rat and human stem cells and provides a foundation for future genome-wide efforts aimed at deciphering the function of all genes encoded by the mammalian genome." | ||
Revision as of 09:53, 26 March 2015
| Embryology - 18 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
The mouse (taxon-mus) has always been a good embryological model, generating easily (litters 8-20) and quickly (21d). Mouse embryology really expanded when molecular biologists used mice for gene knockouts. The term "knockout mouse" is a mouse where researchers have inactivated, or "knocked out," an existing gene by replacing it or disrupting it with an artificial piece of DNA. Currently there is a scientific drive to establish a knockout for all known mouse genes, and new technologies allow both "conditional knockouts" and "knockins".
- Now it is necessary to understand development, in order to understand the effect of knocking out these gene.
--Mark Hill (talk) 08:35, 24 June 2014 (EST) Note the external links below no longer function. The database has been relocated. I will remove this notice when links are repaired.
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
mouse | Molecular Development - Genetics
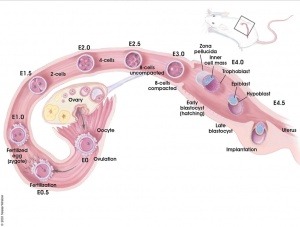
- Mouse Stages: E1 | E2.5 | E3.0 | E3.5 | E4.5 | E5.0 | E5.5 | E6.0 | E7.0 | E7.5 | E8.0 | E8.5 | E9.0 | E9.5 | E10 | E10.5 | E11 | E11.5 | E12 | E12.5 | E13 | E13.5 | E14 | E14.5 | E15 | E15.5 | E16 | E16.5 | E17 | E17.5 | E18 | E18.5 | E19 | E20 | Timeline | About timed pregnancy
| Carnegie | Stage | |||||||||||||||||||||||
| Human | Days | 1 | 2-3 | 4-5 | 5-6 | 7-12 | 13-15 | 15-17 | 17-19 | 20 | 22 | 24 | 28 | 30 | 33 | 36 | 40 | 42 | 44 | 48 | 52 | 54 | 55 | 58 |
| Mouse | Days | 1 | 2 | 3 | E4.5 | E5.0 | E6.0 | E7.0 | E8.0 | E9.0 | E9.5 | E10 | E10.5 | E11 | E11.5 | E12 | E12.5 | E13 | E13.5 | E14 | E14.5 | E15 | E15.5 | E16 |
| Rat | Days | 1 | 3.5 | 4-5 | 5 | 6 | 7.5 | 8.5 | 9 | 10.5 | 11 | 11.5 | 12 | 12.5 | 13 | 13.5 | 14 | 14.5 | 15 | 15.5 | 16 | 16.5 | 17 | 17.5 |
| Note these Carnegie stages are only approximate day timings for average of embryos. Links: Carnegie Stage Comparison | ||||||||||||||||||||||||
| ||||||||||||||||||||||||
| Timeline Links: human timeline | mouse timeline | mouse detailed timeline | chicken timeline | rat timeline | Medaka | Category:Timeline |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Knockout Mouse <pubmed limit=5>Knockout Mouse</pubmed> |
Mouse Gene Knockouts Listed according to the Name of the Gene
List from bioscience.org mouse gene knockouts http://www.bioscience.org/knockout/alphabet.htm
A
- Acid Alpha-glucosidase gene (Gaa)
- Acrosin
- Acute Regulatory Protein (StAR)
- Adenosine A2a receptor
- Adenosine deaminase (ADA)
- alpha 1 (IX) collagen
- alpha calcium-calmodulin kinase II (alpha CaMKII)
- Alpha1b-adrenergic
- Alpha-Galactosidase A
- Apc(delta716)
- Amyloid precursor protein (APP)
- Angiotensin-converting enzyme (ACE)
- Angiotensinogen
- aP2
- ApoA-I
- Apo B
- ApoC-III
- Apo E
- Apolipoprotein A-IV
- Atm
B
- B7
- Bax
- Bc16
- Bcl-2
- Bcl-3
- Bcl-x
- Beta 1, 4-galactosyltransferase (GalTase)
- Beta 1 integrin
- Beta 2-microglobulin
- Beta-3 GABAA receptor
- Beta-galactosidase
- BMP7
- Bradykinin B2 receptor
- Brain-derived neurotrophic factor (BDNF)
C
- C5a receptor
- c-abl
- Calcineurin A alpha
- CD2
- CD22
- CD28
- CD3 epsilon
- CD3 eta/phi
- CD3 zeta/eta
- CD30
- CD4
- CD40
- CD40L
- CD45 and p561ck
- CD45-exon 6 protein tyrosine phosphatase
- CD8
- CD8 beta-1
- C/ebp Alpha
- C/EBP beta (CCAAT/enhancer-binding protein beta)
- Cellular glutathione peroxidase (GSHPX-1)
- c-Fos
- Ciliary Neurotrophic Factor
- c-jun
- C kappa
- c-mos
- c-myc
- CNTF
- CNTFR alpha
- Cox 2
- CRABPI
- Creatine kinase (CK)
- CREB
- CREM
- c-rel proto-oncogene
- c-ret
- CRH
- CSF-1
- CSK
- Ctla-4
- Cystic fibrosis transmembrane conductance regulator (CFTR)
D
- D1A dopamine receptors
- Dazla
- Desmin
- DNA methyl transferase
- DNA repair gene (ERCC-1)
- Dopamine D2 receptors
- Dopamine D4 receptor
- Dv11
E
- E2a
- E2f-1
- E-cadherin
- EDN3
- EGFR
- En-1
- En-2
- Endothelial selectins
- Endothelin-B receptor (EDNRB)
- Enx (Hox 11 L1)
- Epidermal growth factor receptor (Egfr)
- ErbB4
- Estrogen receptor
- Ets-related factor TEL
- Factor IX
- Fas and Perforin
- Fc epsilon RI
- FcR gamma chain
- Fgf4
- FGF5
- FGF-6
- Fibrillin-1
- Fmr1
- FosB
- FSH b subunit
- Ftz-F1
- fyn
- G-CSF
- GAD67
- Gelationase A (matrix metalloproteinase)
- Gelsolin
- Glial fibrillary acidic protein (GFAP)
- Glucocerebrosidase
- GluR-B
- GLUT4
- GM-CSF
- G protein-coupled, inwardly rectifying K+ channel GIRK2
- G protein-coupled receptor kinase 3
- Growth hormone receptor/binding protein
- H2-M
- Hdh ex 5
- Helix-loop-helix gene E2A
- Hoxa-1
- Hoxa-2
- Hoxa-4
- Hoxa9
- Hoxa-11
- Hoxb-4
- Hoxd-3
- Hoxd-13
- Hsp 70-2
- ICAM-1
- ICAM-1 and P selectin
- IFN-gamma
- IFN-gamma receptor
- Ikaros
- IL-1 Beta Converting Enzyme
- IL-1 type I receptor
- IL-2
- IL-2 receptor beta
- IL-4
- IL-6
- IL-7
- IL-10
- IL-18 (IGIF)
- Immunoglobulin D
- Immunoglobulin heavy chain joining region
- Immunoglobulin heavy chain intron enhancer
- Immunoglobulin kappa chain
- Immunoglobulin kappa chain intron enhancer
- Immunoglobulin mu chain
- Inhibin
- Inducible nitric oxide synthase (iNOS)
- int-1
- int-2
- Insulin (Ins1, Ins2)
- Insulin-like growth factor I (Igf-1)
- Insulin-like growth factor I receptor (Igf1r))
- Insulin-like growth factor II (IGF-II)
- Insulin-like growth factor type 2 (Igf2) receptor
- Insulin receptor substrate-1 (IRS-1)
- Integrin-associated protein (IAP)
- Interferon consensus sequence binding protein
- Interferon regulatory factor 1 (IRF-1)
- Interferon regulatory factor 2 (IRF-2)
- Interleukin-2
- ">Interleukin-6 (IL-6)
- Interleukin-7
- Interleukin-7 receptor
- Interleukin-11 receptor (IL11RA)
- IX factor
- L14 S-type lectin
- Lag3
- Lama2
- Lck
- Lecithin: cholestorel acyltransferase (LCAT)
- LIF
- L-isoaspartate (D-aspartate) O-methyltransferase (EC 2.1.1.77)
- Low density lipoprotein (LDL) receptor
- LDL Receptor-Related Protein
- L-selectin
- Lymphotoxin
- Lyn
- Mammalian achaete-scute homolog 1 (Mash-1)
- Mannose 6-phosphate receptor
- mdr2
- mdrla P-glycoprotein
- Melanocortin-4 receptor
- Metallothionein I and II (MT I and MT II)
- Mgat-1
- mGluR1
- mGluR2
- mGluR6
- MHC class I and MHC class II
- MHC class I
- MHC class II
- MHC class II invariant chain
- MKK4
- MLH1
- MLP
- Mu chain
- Mu-opioid receptor
- Mullerian-inhibiting substance (MIS)
- Muscle creatine kinase (M-CK)
- Myf-5 and MyoD
- Myogenin
- N-cadherin
- Neural-cell adhesion molecule (N-CAM)
- Ncx/Hox 11L.1
- Neural nitric oxide synthase (nNOS)
- Neuropeptide Y (NPY)
- Neuregulin
- Neurotrophin-3 (NT3)
- Neurotrophin-4 (NT4)
- Nuetral Endopeptidase
- Nf1
- NFAT1
- NF-kappaB2
- NGF
- NGFI-A (Egr-1)(Krox-24)
- NMDA receptor 1 (NMDAR1)
- N-myc
- p21 CIP1/WAF1
- p27kip1
- p53
- p561ck and CD45
- P75NTR
- PDE gamma
- Pemphigus vulgaris antigen (desmoglein 3; dsg3)
- Perforin
- Perforin and Fas
- Peripheral myelin protein 22 (PM22)
- Phosphatidylethanolamine N-methyltransferase
- PKC beta
- PKCgamma
- Pkd1
- Plasminogen activator (PA)
- Plasminogen activator inhibitor-1 (PAI-1)
- Platelet-derived growth factor (PDGF)
- Plp
- PO
- Poly(ADP-ribose) polymerase
- Polycomb-M33
- Presenlin-1
- Pre-proenkephalin
- Progesterone receptor (PR)
- Proteasome subunit LMP-7
- PrP
- P selectin
- P and E selectins
- P selectin and Intercellular adhesion molecule 1 (ICAM-1)
- Pxr1
- R II alpha
- RII beta subunit of Protein Kinase A
- Rad51
- rag-2
- Rb-1
- Retinoic acid receptor alpha (RAR alpha)
- RelB
- Renin Ren-1d
- Rhodopsin
- Rod cGMP phosphodiesterase gamma subunit
- RXR alpha
- Selenocysteine tRNA gene (Trsp) -
- Semaphorin III/D
- SMN
- SP-B
- Sp1
- Stat 1
- STAT5A
- SREBP-1
- Synapsin I
- TAP1
- Tensin
- TCR-alpha-beta
- TCR delta
- T cell receptor zeta chain
- TdT
- Tenascin C
- TGF-alpha
- TGF-beta 1
- TGFbeta2
- TIMP-1
- Tissue factor (TF)
- TNF receptor I
- t-PA
- TPO
- Transthyretin
- trkB
- Tumor necrosis factor
- Tyrosine Phosphatase
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- International Knockout Mouse Consortium (IKMC) The International Knockout Mouse Consortium (IKMC) aims to mutate all protein-coding genes in the mouse using gene trapping and gene targeting in C57BL/6 ES cells.
- NIH - The Knockout Mouse Project (KOMP) Is a trans-National Institutes of Health (NIH) initiative that aims to generate a comprehensive and public
- KOMP The University of California, Davis (UC Davis) and Children's Hospital Oakland Research Institute (CHORI) will collaborate to preserve, protect and make available knockout mice and related products available to the research community. The repository will archive, maintain and distribute up to 8,500 strains of embryonic stem cell clones, live mouse lines, frozen embryos and sperm and vectors.
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 18) Embryology Mouse Knockout. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Mouse_Knockout
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G
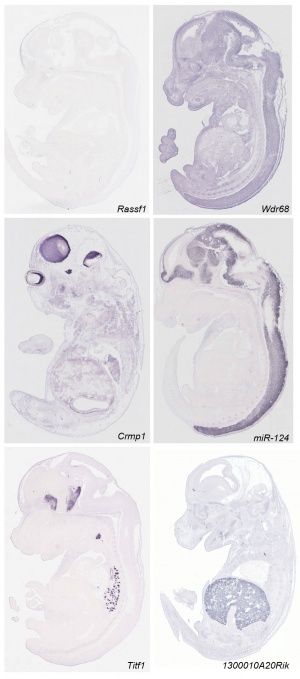
- ↑ <pubmed>21267068</pubmed>| PLoS Biol. | Eurexpress transcriptome atlas
- ↑ <pubmed>25803277</pubmed>
- ↑ <pubmed>21677718</pubmed>| Nature News
- ↑ <pubmed>21677750</pubmed>