Mouse Development: Difference between revisions
| Line 260: | Line 260: | ||
{{External Links}} | {{External Links}} | ||
* Mouse Gene Expression | ===Mouse Embryology=== | ||
* [http://www.cshl.org/books/manipulating_mouse_embryo.html Manipulating the Mouse Embryo (Cold Spring Harbor Laboratory)] | |||
* [http://embryo.mc.duke.edu/animal/cdAtlas/cdAtlas.html Digital Atlas of Mouse Embryology] | |||
===Mouse Gene Expression=== | |||
* [http://www.emouseatlas.org/emage/ EMAGE] is a database of in situ gene expression data in the mouse embryo and an accompanying suite of tools to search and analyse the data. (All site content, except where otherwise noted, is licensed under a [http://creativecommons.org/licenses/by/3.0/ Creative Commons Attribution License].) | |||
* [http://www.eurexpress.org Eurexpress transcriptome atlas] a High-Resolution Anatomical Atlas of the Transcriptome in the Mouse Embryo. | |||
* [http://genepaint.org/Frameset.html GenePaint.org] is a digital atlas of gene expression patterns in the mouse. | |||
===Mouse Genome=== | |||
* [http://www.ncbi.nlm.nih.gov/genome/seq/MmHome.html NCBI- Mouse Genome Sequencing] | * [http://www.ncbi.nlm.nih.gov/genome/seq/MmHome.html NCBI- Mouse Genome Sequencing] | ||
* [http://www.informatics.jax.org/ Mouse Genome Informatics] | * [http://www.informatics.jax.org/ Mouse Genome Informatics] | ||
* [http://www.ebi.ac.uk/genomes/mot/10090_1000.html EBI- Mouse Genome Sequencing] | * [http://www.ebi.ac.uk/genomes/mot/10090_1000.html EBI- Mouse Genome Sequencing] | ||
* [http://www.anex.med.tokushima-u.ac.jp/index.html ANEX Rat and Mouse Database - (University of Tokushima)] | * [http://www.anex.med.tokushima-u.ac.jp/index.html ANEX Rat and Mouse Database - (University of Tokushima)] | ||
* [http://www.genome.wi.mit.edu/cgi-bin/mouse/index Genetic and Physical Maps of the Mouse Genome (Whitehead Institute/MIT)] | |||
* [http://cedar.genetics.soton.ac.uk/public_html/ Genetic Location Database (University of Southampton)] | |||
* [http://darwin.ceh.uvic.ca/people/koop/KoopGen.html Koop Human and Mouse Genome Project Research (University of Victoria)] | |||
* [http://www.genomesystems.com/ Genome Systems, Inc.] | |||
===Mouse Jackson Laboratory=== | |||
* [http://www.jax.org/ Jackson Laboratory] | |||
* [http://www.jax.org/resources/documents/imr/ Jackson Laboratory Induced Mutant Resource] | |||
* [http://www.informatics.jax.org/doc/lists.html Jackson Laboratory BioInformatics Electronic Bulletin Board System] | |||
===Mouse Diseases=== | |||
* [http://www.cdc.gov/ncidod/EID/vol1no4/rice2.htm Helicobacter hepaticus, a Recently Recognized Bacterial Pathogen, Associated with Chronic Hepatitis and Hepatocellular Neoplasia in Laboratory Mice (CDC)] | |||
* [http://www.criver.com/techdocs/helico-1.html Helicobacter Infection in Laboratory Mice: History, Significance, Detection and Management (Charles River)] | |||
* [http://www.nap.edu/bookstore/isbn/0309037948.html Infectious Diseases of Mice and Rats (NAS-ILAR)] | |||
* [http://www.nap.edu/bookstore/isbn/0309042836.html Infectious Diseases of Mice and Rats Companion Guide (NAS-ILAR)] | |||
* [http://www.afip.org/vetpath/POLA/micerat.txt Infectious Diseases of Mice and Rats (AFIP-POLA)] | |||
* [http://www.ncifcrf.gov/VETPATH/emerg.html Murine Helicobacters (NCI)] | |||
===Mouse Transgenics=== | |||
* [http://www.lists.ic.ac.uk/hypermail/transgenic-list/ Archives of Transgenic-List] | * [http://www.lists.ic.ac.uk/hypermail/transgenic-list/ Archives of Transgenic-List] | ||
* [http://condor.mbcr.bcm.tmc.edu/BEP/ERMB/home.html Mammary Transgene Database (Baylor College of Medicine)] | |||
* [http://www.cshl.org/books/targeted_mutagenesis.html Targeted Mutagenesis in Mice: A Video Guide] | |||
* [http://www.bis.med.jhmi.edu/Dan/tbase/tbase.html TBASE - Transgenic/Targeted Mutation Database] | |||
* [http://www.bku.com/ B Universal Ltd.] | * [http://www.bku.com/ B Universal Ltd.] | ||
* [http://www.bio.net/hypermail/BIGBLUE/ BIG BLUE (Big Blue Transgenic Mouse Mailing List Archives)] | * [http://www.bio.net/hypermail/BIGBLUE/ BIG BLUE (Big Blue Transgenic Mouse Mailing List Archives)] | ||
| Line 274: | Line 303: | ||
* [http://www.criver.com/ Charles River Laboratories, Inc.] | * [http://www.criver.com/ Charles River Laboratories, Inc.] | ||
* [http://www.chrysalis-cro.com/transgenic/ Chrysalis DNX Transgenic Services] | * [http://www.chrysalis-cro.com/transgenic/ Chrysalis DNX Transgenic Services] | ||
* [http://www. | * [http://www.ucc.uconn.edu/%7Ewwwbiotc/Transg.html Transgenic Animal Facility (University of Connecticut Biotechnology Center)] | ||
* [http:// | * [http://www.life.uiuc.edu/biotech/transgenic.html Transgenic Animal Facility (University of Illinois Biotechnology Center)] | ||
* [http://www.uiowa.edu/~vpr/research/units/transgen.htm Transgenic Animal Facility (University of Iowa)] | |||
* [http://www.ki.se/core/meg.html Transgenic Animal Facility (Karolinska Institute)] | |||
* [http://ccmb.microbiol.uwa.edu.au/ccmbtrans.html Transgenic Animal Facility (University of Western Australia)] | |||
* [http://www.biotech.wisc.edu/Service/transan.html Transgenic Animal Facility (University of Wisconsin Biotechnology Center)] | |||
* [http://www.med.umich.edu/tamc/ Transgenic Animal Model Core (University of Michigan)] | |||
* [http://www.bcm.tmc.edu/urol/spore/transgen_core.html Transgenic Core (Baylor College of Medicine SPORE)] | |||
* [http://www.uchsc.edu/sm/animal/tmcl/txhmpg.html Transgenic Facility (University of Colorado Health Science Center)] | |||
* [http://www.uky.edu/Transgenic/ Transgenic Facility (University of Kentucky)] | |||
* [http://som1.ab.umd.edu/Microbiology/Transgene.html Transgenic & Knockout Mouse Facility (University of Maryland at Baltimore)] | |||
* [http://www.lists.ic.ac.uk/hypermail/transgenic-list/ Transgenic-List Archives] | |||
* [http://www-rics.bwh.harvard.edu/derm/tg_core.html Transgenic Models of Skin Disease Core (Harvard Skin Disease Research Center)] | |||
* [http://www.mssm.edu/molbio/bcmb/BCMB_Core_Facilities.html Transgenic Mouse Core Facilities (Mount Sinai School of Medicine)] | |||
* [http://www.med.virginia.edu/~sp3i/tghome.html Transgenic Mouse Core Facility (University of Virginia)] | |||
* [http://cpmcnet.columbia.edu/research/equip/eq-cctrn.htm Transgenic Mouse Facility (Columbia-Presbyterian Cancer Center)] | |||
* [http://genetics.mc.duke.edu/transgenX.html Transgenic Mouse Facility (Duke Comprehensive Cancer Center)] | |||
* [http://www.pharm.sunysb.edu/transgenic/Transgenic.html Transgenic Mouse Facility (SUNY-Stony Brook)] | |||
* [http://darwin.bio.uci.edu/~tjf/index.html Transgenic Mouse Facility (University of California-Irvine)] | |||
* [http://www.cosm.sc.edu/ibrt/mouse.html Transgenic Mouse Facility (University of South Carolina)] | |||
* [http://info.med.yale.edu/ycc/transgenic.html Transgenic Mouse Facility (Yale University Cancer Center)] | |||
* [http://www-mp.ucdavis.edu/tgmice/firststop.html Transgenic Mouse Program (University of California-Davis)] | |||
* [http://kccc-www.med.nyu.edu/mouse.htm Transgenic Mouse Research Facility (New York University Kaplan Comprehensive Cancer Center)] | |||
* [http://www.transgenics-berlin.com/ Transgenics (Berlin-Buch GmbH)] | |||
===Mouse Unsorted Links=== | |||
* [http://www.covance.com/ Covance Research Products] | |||
* [http://www.cns.ohiou.edu/ictto/ebi/transgenic.html Edison Biotechnology Institute (Ohio University)] | * [http://www.cns.ohiou.edu/ictto/ebi/transgenic.html Edison Biotechnology Institute (Ohio University)] | ||
* [http://www.informatics.jax.org/encyclo.html Encyclopedia of the Mouse Genome (Jackson Laboratory)] | * [http://www.informatics.jax.org/encyclo.html Encyclopedia of the Mouse Genome (Jackson Laboratory)] | ||
* [http://www.hgmp.mrc.ac.uk/MBx/MBxHomepage.html EUCIB Mouse Backcross Database (MBx)] | * [http://www.hgmp.mrc.ac.uk/MBx/MBxHomepage.html EUCIB Mouse Backcross Database (MBx)] | ||
* [http://www.harlan.com/ Harlan Sprague Dawley, Inc.] | * [http://www.harlan.com/ Harlan Sprague Dawley, Inc.] | ||
* [http://www.bis.med.jhmi.edu/Dan/tbase/docs/knockout.html It's a Knockout] | * [http://www.bis.med.jhmi.edu/Dan/tbase/docs/knockout.html It's a Knockout] | ||
* [gopher://hobbes.jax.org:70/11/ Jackson Laboratory Gopher Server] | * [gopher://hobbes.jax.org:70/11/ Jackson Laboratory Gopher Server] | ||
* [gopher://hobbes.informatics.jax.org:70/00/pub/stocks/lanelist.txt Lane List of Named Mutations and Polymorphic Loci] | * [gopher://hobbes.informatics.jax.org:70/00/pub/stocks/lanelist.txt Lane List of Named Mutations and Polymorphic Loci] | ||
* [http://www.lexgen.com/ Lexicon Genetics, Inc.] | * [http://www.lexgen.com/ Lexicon Genetics, Inc.] | ||
* [http://www.cdc.gov/ncidod/EID/vol1no4/barton.htm#top Lymphocytic Choriomeningitis Virus: An Unrecognized Teratogenic Pathogen (CDC)] | * [http://www.cdc.gov/ncidod/EID/vol1no4/barton.htm#top Lymphocytic Choriomeningitis Virus: An Unrecognized Teratogenic Pathogen (CDC)] | ||
* [http://www.m-b.dk/ M & B Breeding and Research Centre Ltd.] | * [http://www.m-b.dk/ M & B Breeding and Research Centre Ltd.] | ||
* [http://mcbio.med.buffalo.edu/mapmgr.html Map Manager Software] | * [http://mcbio.med.buffalo.edu/mapmgr.html Map Manager Software] | ||
* [http://www.ornl.gov/ORNLReview/rev27-12/text/mnmmain.html Mice and Men: Making the Most of Our Similarities (ORNL)] | * [http://www.ornl.gov/ORNLReview/rev27-12/text/mnmmain.html Mice and Men: Making the Most of Our Similarities (ORNL)] | ||
| Line 305: | Line 345: | ||
* [http://genex.hgu.mrc.ac.uk/ Mouse Atlas and Gene Expression Database Project] | * [http://genex.hgu.mrc.ac.uk/ Mouse Atlas and Gene Expression Database Project] | ||
* [http://ws4.niai.affrc.go.jp/dbsearch2/mmap/mmap.html Mouse Cytogenetic Map Image] | * [http://ws4.niai.affrc.go.jp/dbsearch2/mmap/mmap.html Mouse Cytogenetic Map Image] | ||
* [gopher://larry.pathology.washington.edu:70/11/idiograms/Mouse Mouse Idiograms] | * [gopher://larry.pathology.washington.edu:70/11/idiograms/Mouse Mouse Idiograms] | ||
* [http://www.informatics.jax.org/bin/strains/search Mouse Inbred Strains (Jackson Labs)] | * [http://www.informatics.jax.org/bin/strains/search Mouse Inbred Strains (Jackson Labs)] | ||
| Line 313: | Line 352: | ||
* [http://hyperarchive.lcs.mit.edu/HyperArchive/Archive/sci/mouse-up-15-hc.hqx MouseUp HyperCard Transgenic Mouse Database (Macintosh)] | * [http://hyperarchive.lcs.mit.edu/HyperArchive/Archive/sci/mouse-up-15-hc.hqx MouseUp HyperCard Transgenic Mouse Database (Macintosh)] | ||
* [http://www.mgc.har.mrc.ac.uk/ MRC Mouse Genome Centre] | * [http://www.mgc.har.mrc.ac.uk/ MRC Mouse Genome Centre] | ||
* [http://www.grs.nig.ac.jp/NIG_mouse/mouse.default.html National Institute of Genetics Mouse Genetic Resources (Japan)] | * [http://www.grs.nig.ac.jp/NIG_mouse/mouse.default.html National Institute of Genetics Mouse Genetic Resources (Japan)] | ||
* [http://www.nih.gov/od/ors/dirs/vrp/nihagr.htm NIH Animal Genetic Resource] | * [http://www.nih.gov/od/ors/dirs/vrp/nihagr.htm NIH Animal Genetic Resource] | ||
| Line 324: | Line 362: | ||
* [http://www.criver.com/techdocs/past-1.html Review of Pasteurella pneumotropica (Charles River)] | * [http://www.criver.com/techdocs/past-1.html Review of Pasteurella pneumotropica (Charles River)] | ||
* [http://www.taconic.com/ Taconic] | * [http://www.taconic.com/ Taconic] | ||
* [http://www.vetmed.ucdavis.edu/Animal_Alternatives/cancer.htm The Mouse in Science: Cancer Research] | * [http://www.vetmed.ucdavis.edu/Animal_Alternatives/cancer.htm The Mouse in Science: Cancer Research] | ||
* [http://www.vetmed.ucdavis.edu/Animal_Alternatives/mabs.htm The Mouse in Science: Monoclonal Antibodies] | * [http://www.vetmed.ucdavis.edu/Animal_Alternatives/mabs.htm The Mouse in Science: Monoclonal Antibodies] | ||
* [http://www.vetmed.ucdavis.edu/Animal_Alternatives/vaccines.htm The Mouse in Science: Vaccines] | * [http://www.vetmed.ucdavis.edu/Animal_Alternatives/vaccines.htm The Mouse in Science: Vaccines] | ||
* [http://www.vetmed.ucdavis.edu/Animal_Alternatives/whymice.htm The Mouse in Science: Why Mice?] | * [http://www.vetmed.ucdavis.edu/Animal_Alternatives/whymice.htm The Mouse in Science: Why Mice?] | ||
Revision as of 15:31, 26 May 2011
Introduction
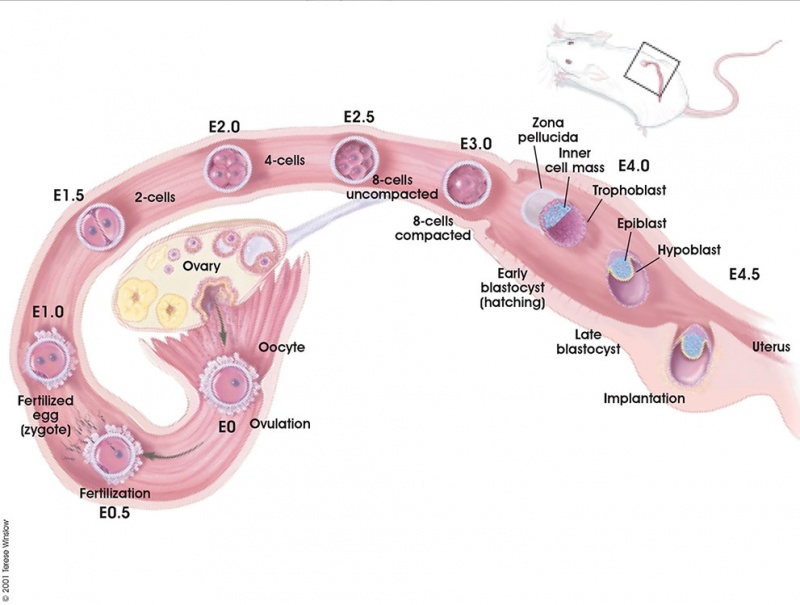
The mouse (taxon-mus) has always been a good embryological model, generating easily (litters 8-20) and quickly (21d). Mouse embryology really expanded when molecular biologists used mice for gene knockouts. Suddenly it was necessary to understand development in order to understand the effect of knocking out the gene. There are over 450 different strains of inbred research mice, and these strains have recently been organized into a chart. While being an ideal model organism, only a relatively small amount (1.5%) of the total mouse genome has been sequenced. Those interested in the mouse reproductive cycle should also look at the mouse estrous cycle.
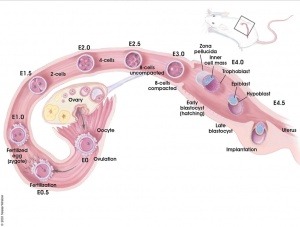
There are several systems for staging mouse development. The original and most widely used is the Theiler Stages system, which divides mouse development into 26 prenatal and 2 postnatal stages. [1]
- Mouse Stages: E1 | E2.5 | E3.0 | E3.5 | E4.5 | E5.0 | E5.5 | E6.0 | E7.0 | E7.5 | E8.0 | E8.5 | E9.0 | E9.5 | E10 | E10.5 | E11 | E11.5 | E12 | E12.5 | E13 | E13.5 | E14 | E14.5 | E15 | E15.5 | E16 | E16.5 | E17 | E17.5 | E18 | E18.5 | E19 | E20 | Timeline | About timed pregnancy
| Carnegie | Stage | |||||||||||||||||||||||
| Human | Days | 1 | 2-3 | 4-5 | 5-6 | 7-12 | 13-15 | 15-17 | 17-19 | 20 | 22 | 24 | 28 | 30 | 33 | 36 | 40 | 42 | 44 | 48 | 52 | 54 | 55 | 58 |
| Mouse | Days | 1 | 2 | 3 | E4.5 | E5.0 | E6.0 | E7.0 | E8.0 | E9.0 | E9.5 | E10 | E10.5 | E11 | E11.5 | E12 | E12.5 | E13 | E13.5 | E14 | E14.5 | E15 | E15.5 | E16 |
| Rat | Days | 1 | 3.5 | 4-5 | 5 | 6 | 7.5 | 8.5 | 9 | 10.5 | 11 | 11.5 | 12 | 12.5 | 13 | 13.5 | 14 | 14.5 | 15 | 15.5 | 16 | 16.5 | 17 | 17.5 |
| Note these Carnegie stages are only approximate day timings for average of embryos. Links: Carnegie Stage Comparison | ||||||||||||||||||||||||
| ||||||||||||||||||||||||
| Timeline Links: human timeline | mouse timeline | mouse detailed timeline | chicken timeline | rat timeline | Medaka | Category:Timeline |
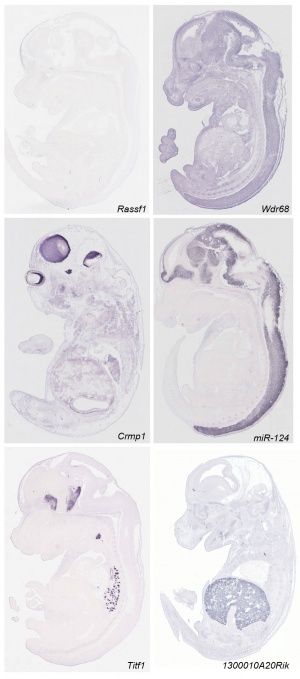
Some Recent Findings
|
Early Mouse Development
- Links: Mouse Stages
Later Mouse Development
Mouse E8.5[3]
Mouse E9.5[4]
Mouse E10.5[4]
Mouse E11.5[4]
Mouse E11.5[4]
Mouse E12.5[4]
Mouse E14.5[5]
- Links: Mouse Stages | Mouse Timeline Detailed
Carnegie Stages Comparison
The table below gives an approximate comparison of human, mouse and rat embryos based upon Carnegie staging.
| Species | Stage | |||||||||||||||
| Human [6] | Days | 20 | 22 | 24 | 28 | 30 | 33 | 36 | 40 | 42 | 44 | 48 | 52 | 54 | 55 | 58 |
| Mouse [1] | Days | 9 | 9.5 | 10 | 10.5 | 11 | 11.5 | 12 | 12.5 | 13 | 13.5 | 14 | 14.5 | 15 | 15.5 | 16 |
| Rat [7] | Days | 10.5 | 11 | 11.5 | 12 | 12.5 | 13 | 13.5 | 14 | 14.5 | 15 | 15.5 | 16 | 16.5 | 17 | 17.5 |
Spermatozoa Development
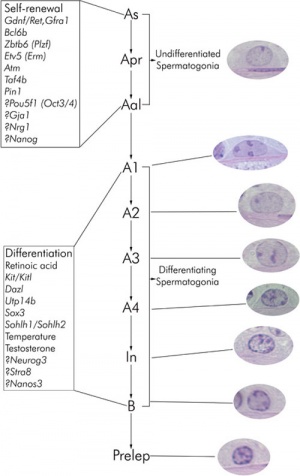
The process of spermatogenesis takes approximately 35 days:
- mitotic phase (11 days)
- meiotic phase (10 days)
- post-meiotic phase (14 days)
Spermatogonial stem cells (SSCs)
The diploid germ cells, spermatogonial stem cells (SSCs), are located on the basement membrane of the seminiferous tubules
- adult mouse testis about 30,000 SSCs
- either divides into two new single cells
- or into a pair of spermatogonia (Apr)
- that do not complete cytokinesis and stay connected by an intercellular bridge
Primitive spermatogonia subset
- Asingle (As, single isolated spermatogonia)
- Apaired (Apr, interconnected spermatogonial pairs)
- Aaligned (Aal, interconnected 4, 8, or 16 spermatogonia)
- specifically termed Aal-4, Aal-8, and Aal-16
Primitive spermatogonia cells transform without cell division into more differentiating A1 spermatogonia that undergo 6 mitotic and 2 meiotic divisions to eventually form haploid spermatids.
Neural Development
The data below is summarised from a study of early neural development in the mouse.[9]
- initial fusion of apposing neural folds occurred at the level of the intermediate point between the third and fourth somites (caudal myelencephalon) both rostrally and caudally
- second fusion - at the original rostral end of the neural plate (rostrodorsally).
- third fusion - in the caudal diencephalon (rostrally and caudally)
- followed by complete closure of the telencephalic neuropore at the midpoint of the telencephalic roof
- then complete closure of the metencephalic neuropore at the rostral part of the metencephalic roof
- fourth fusion - at the original caudal end of the neural plate (rostrally)
- caudal neuropore completely closed at the level of the future 33rd somite
See also these 1980's papers.[10] [11]
Mouse Knockouts
Knowledge about mouse development has rapidly expanded as it has become the model animal system for genetic "knock out " studies. This technology actually requires development of defined breeding programs, pseudo-pregnancy, in vitro fertilization, molecular biology, and good old fashioned histology. Without understanding normal development the molecular biologists don't stand a hope of understanding what their gene knock out has done. There is a database of all existing mouse knockouts and their consequences.
Murine Development Control Genes
Kessel, M. and Gruss, P. Science 249 374-379 (1990)
An early review of the genes, and method of identifying them, involved in early mouse development. In particular discusses Homeobox genes. (homeobox is 183bp encoding a 61 amino acid DNA-binding domain)
- Gene families
- Hox
- Pax
- POU
The Genealogy Chart of Inbred Strains
This chart shows the origins and relationships of inbred mouse strains. The chart is available as a PDF document [../pdf/mouse_genealogy.pdf Locally] or from JAX Labs and was originally published by Beck etal., 2000.
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, Fisher EM. Genealogies of mouse inbred strains. Nat Genet. 2000 Jan;24(1):23-5.
Mouse Genome
Mouse Genome completed December 2002, a draft sequence and analysis of the genome of the C57BL/6J mouse strain.
- less than 30,000 genes
- estimated size is 2.5 Gb, smaller than the human genome
- about 40% of the human and mouse genomes can be directly aligned
- about 80% of human genes have one corresponding gene in the mouse genome
- Links: Mouse Genome Sequencing: Mus musculus | Mouse Genome Informatics | Mouse Genome Project | Nature - Mouse Genome
References
- ↑ 1.0 1.1 The House Mouse: Atlas of Mouse Development by Theiler Springer-Verlag, NY (1972, 1989). | online book
- ↑ 2.0 2.1 <pubmed>21267068</pubmed>| PLoS Biol. | Eurexpress transcriptome atlas
- ↑ <pubmed>20704721</pubmed>| BMC Dev Biol.
- ↑ 4.0 4.1 4.2 4.3 4.4 <pubmed>16683035</pubmed>| PLoS Genetics
- ↑ <pubmed>18713865</pubmed>| PNAS
- ↑ <pubmed>400868</pubmed>
- ↑ Witschi, E. (1962) Development: Rat. In: Growth Including Reproduction and Morphological Development. Altman, P. L. , and D. S. Dittmer, ed. Fed. Am. Soc. Exp. Biol., Washington DC, pp. 304-314.
- ↑ Zhou Q, Griswold MD. Regulation of spermatogonia. StemBook [Internet]. Cambridge (MA): Harvard Stem Cell Institute. PMID20614596 | http://www.ncbi.nlm.nih.gov/books/NBK27035/ Bookshelf]
- ↑ Neurulation in the mouse: manner and timing of neural tube closure. Sakai Y. Anat Rec. 1989 Feb;223(2):194-203. PMID: 2712345
- ↑ The histogenetic potential of neural plate cells of early-somite-stage mouse embryos. Chan WY, Tam PP. J Embryol Exp Morphol. 1986 Jul;96:183-93. PMID: 3805982
- ↑ Neurulation in the mouse. I. The ontogenesis of neural segments and the determination of topographical regions in a central nervous system. Sakai Y. Anat Rec. 1987 Aug;218(4):450-7. PMID: 3662046
Search Pubmed
Search Pubmed: Mouse Development | Mouse Embryology
Additional Images
Movies
| Fertilization mouse |
Parental genome mouse |
Nodal cilia rotation mouse |
Somitogenesis mouse |
| Migration 1 | Migration 2 | Migration 3 | microCT E11.5 |
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
Mouse Embryology
Mouse Gene Expression
- EMAGE is a database of in situ gene expression data in the mouse embryo and an accompanying suite of tools to search and analyse the data. (All site content, except where otherwise noted, is licensed under a Creative Commons Attribution License.)
- Eurexpress transcriptome atlas a High-Resolution Anatomical Atlas of the Transcriptome in the Mouse Embryo.
- GenePaint.org is a digital atlas of gene expression patterns in the mouse.
Mouse Genome
- NCBI- Mouse Genome Sequencing
- Mouse Genome Informatics
- EBI- Mouse Genome Sequencing
- ANEX Rat and Mouse Database - (University of Tokushima)
- Genetic and Physical Maps of the Mouse Genome (Whitehead Institute/MIT)
- Genetic Location Database (University of Southampton)
- Koop Human and Mouse Genome Project Research (University of Victoria)
- Genome Systems, Inc.
Mouse Jackson Laboratory
- Jackson Laboratory
- Jackson Laboratory Induced Mutant Resource
- Jackson Laboratory BioInformatics Electronic Bulletin Board System
Mouse Diseases
- Helicobacter hepaticus, a Recently Recognized Bacterial Pathogen, Associated with Chronic Hepatitis and Hepatocellular Neoplasia in Laboratory Mice (CDC)
- Helicobacter Infection in Laboratory Mice: History, Significance, Detection and Management (Charles River)
- Infectious Diseases of Mice and Rats (NAS-ILAR)
- Infectious Diseases of Mice and Rats Companion Guide (NAS-ILAR)
- Infectious Diseases of Mice and Rats (AFIP-POLA)
- Murine Helicobacters (NCI)
Mouse Transgenics
- Archives of Transgenic-List
- Mammary Transgene Database (Baylor College of Medicine)
- Targeted Mutagenesis in Mice: A Video Guide
- TBASE - Transgenic/Targeted Mutation Database
- B Universal Ltd.
- BIG BLUE (Big Blue Transgenic Mouse Mailing List Archives)
- Big Blue lacI Transgenic Mouse Page
- Charles River Laboratories, Inc.
- Chrysalis DNX Transgenic Services
- Transgenic Animal Facility (University of Connecticut Biotechnology Center)
- Transgenic Animal Facility (University of Illinois Biotechnology Center)
- Transgenic Animal Facility (University of Iowa)
- Transgenic Animal Facility (Karolinska Institute)
- Transgenic Animal Facility (University of Western Australia)
- Transgenic Animal Facility (University of Wisconsin Biotechnology Center)
- Transgenic Animal Model Core (University of Michigan)
- Transgenic Core (Baylor College of Medicine SPORE)
- Transgenic Facility (University of Colorado Health Science Center)
- Transgenic Facility (University of Kentucky)
- Transgenic & Knockout Mouse Facility (University of Maryland at Baltimore)
- Transgenic-List Archives
- Transgenic Models of Skin Disease Core (Harvard Skin Disease Research Center)
- Transgenic Mouse Core Facilities (Mount Sinai School of Medicine)
- Transgenic Mouse Core Facility (University of Virginia)
- Transgenic Mouse Facility (Columbia-Presbyterian Cancer Center)
- Transgenic Mouse Facility (Duke Comprehensive Cancer Center)
- Transgenic Mouse Facility (SUNY-Stony Brook)
- Transgenic Mouse Facility (University of California-Irvine)
- Transgenic Mouse Facility (University of South Carolina)
- Transgenic Mouse Facility (Yale University Cancer Center)
- Transgenic Mouse Program (University of California-Davis)
- Transgenic Mouse Research Facility (New York University Kaplan Comprehensive Cancer Center)
- Transgenics (Berlin-Buch GmbH)
Mouse Unsorted Links
- Covance Research Products
- Edison Biotechnology Institute (Ohio University)
- Encyclopedia of the Mouse Genome (Jackson Laboratory)
- EUCIB Mouse Backcross Database (MBx)
- Harlan Sprague Dawley, Inc.
- It's a Knockout
- Jackson Laboratory Gopher Server
- Lane List of Named Mutations and Polymorphic Loci
- Lexicon Genetics, Inc.
- Lymphocytic Choriomeningitis Virus: An Unrecognized Teratogenic Pathogen (CDC)
- M & B Breeding and Research Centre Ltd.
- Map Manager Software
- Mice and Men: Making the Most of Our Similarities (ORNL)
- Microinjection Workshop
- Mouse Atlas and Gene Expression Database Project
- Mouse Cytogenetic Map Image
- Mouse Idiograms
- Mouse Inbred Strains (Jackson Labs)
- Mouse Linkage Map
- Mouse Nomenclature Rules and Guidelines (Jackson Labs)
- Mouse and Rat Research Home Page
- MouseUp HyperCard Transgenic Mouse Database (Macintosh)
- MRC Mouse Genome Centre
- National Institute of Genetics Mouse Genetic Resources (Japan)
- NIH Animal Genetic Resource
- NIH Directory of Transgenic Mice
- NIH Mouse Club
- Of Mice and Men (HHMI's Blazing a Genetic Trail)
- Online Mendelian Inheritance in Animals (University of Sydney)
- Pathology of Genetically-altered Mice
- Portable Dictionary of the Mouse Genome
- Review of Pasteurella pneumotropica (Charles River)
- Taconic
- The Mouse in Science: Cancer Research
- The Mouse in Science: Monoclonal Antibodies
- The Mouse in Science: Vaccines
- The Mouse in Science: Why Mice?
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 20) Embryology Mouse Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Mouse_Development
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G


![Mouse E8.5[3]](/embryology/images/thumb/9/9f/Mouse-E8.5_dorsal_view.jpg/120px-Mouse-E8.5_dorsal_view.jpg)
![Mouse E9.5[4]](/embryology/images/thumb/b/b2/Mouse-E9.5.jpg/90px-Mouse-E9.5.jpg)
![Mouse E10.5[4]](/embryology/images/thumb/8/89/Mouse_CT_E10.5.jpg/90px-Mouse_CT_E10.5.jpg)
![Mouse E11.5[4]](/embryology/images/thumb/b/b2/Mouse_CT_E11.5.jpg/90px-Mouse_CT_E11.5.jpg)
![Mouse E12.5[4]](/embryology/images/thumb/d/da/Mouse_CT_E12.5.jpg/90px-Mouse_CT_E12.5.jpg)
![Mouse E14.5[5]](/embryology/images/thumb/f/fe/Mouse_CT_E14.5.jpg/77px-Mouse_CT_E14.5.jpg)