Morula Development
| Embryology - 16 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
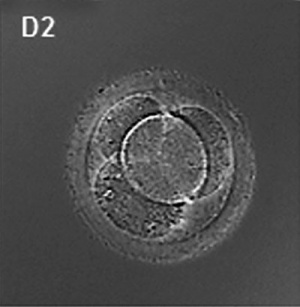
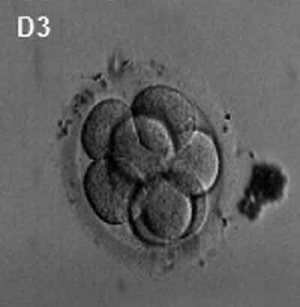
(Latin, morula = mulberry) An early stage in post-fertilization development when cells have rapidly mitotically divided to produce a solid mass of cells (12-15 cells) with a "mulberry" appearance. This stage is followed by formation of a cavity in this cellular mass blastocyst stage.
A key event prior to morula formation is "compaction", where the 8 cell embryo undergoes changes in cell morphology and cell-cell adhesion that initiates the formation of this solid ball of cells.
In humans, morula stage of development occurs during the first days in week one following fertilization (GA week 3) and is described as Carnegie stage 2. This stage is followed by formation of a cavity, the blastocoel, which defines formation of the blastocyst.
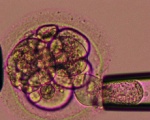 ART Preimplantation blastomere biopsy[2]
|
In Assisted Reproductive Technology, the morula stage is when one of the earliest prenatal diagnostic test can be carried out, by removing a single cell (blastomere) and carrying out genetic diagnosis on its DNA. |
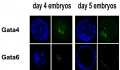
Molecular - In the mouse, during transition from morula to blastocyst stage, differentiation of the inner cell mass (ICM) and trophectoderm (TE) has been shown to be regulated by the Hippo pathway.[3][4]
| Morula Links: morula | Carnegie stage 2 | mitosis | blastocyst | fertilization | Week 1 | Lecture - Week 1 | Category:Carnegie Stage 2 | Category:Morula |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Morula Development |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Movies
|
|
|
|
|
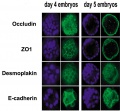
Compaction
- E-cadherin mediated adhesion initiates at compaction at the 8-cell stage
- regulated post-translationally via protein kinase C and other signalling molecules
Blastomere Division
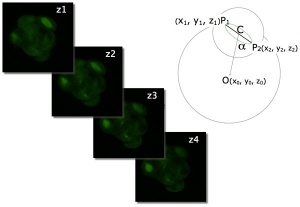
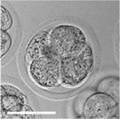
An in vitro study of human blastocyst development[9] showed that those blastomeres that initially divide quickly are more likely to develop to blastocyst stage.
A recent study in mice showed that there was no specific orientation of the mitotic spindle during cell division in the 8 to 16 cell stage transition.[8] This suggests no predetermined cleavage pattern (pre-patterned) at the 8 cell stage and only modulated by the extent of cell rounding up during mitosis. In other species, such as the worm C.elegans and ascidians, have specific patterns of spindle orientation from the zygote stage.
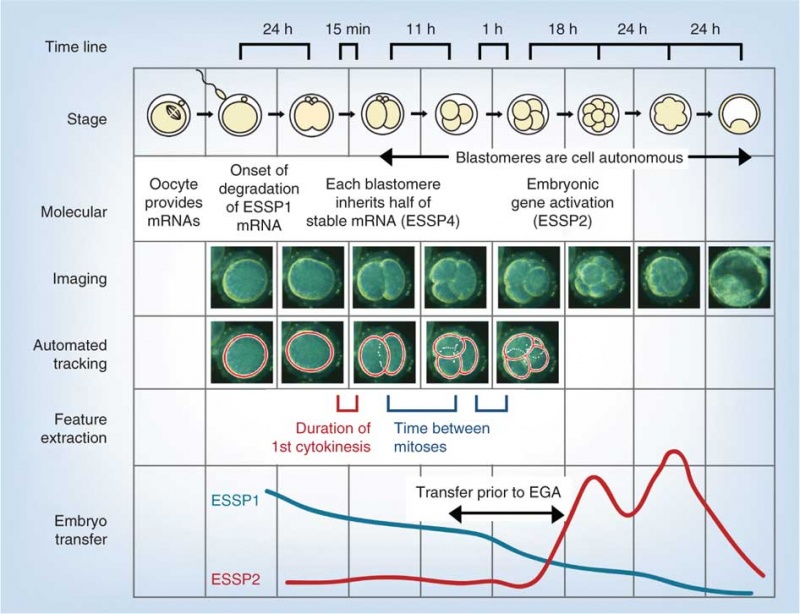
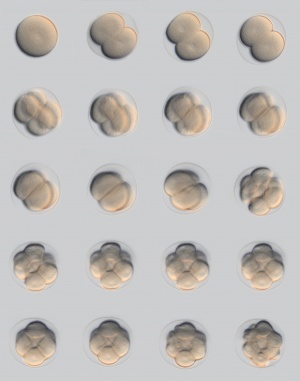
Model Human Morula Development
The following figure is from a recent study[6] using video and genetic analysis of in vitro human development during week 1 following fertilization.
- EGA - embryonic genome activation
- ESSP - embryonic stage–specific pattern, four unique embryonic stage–specific patterns (1-4)
- Links: Figure with legend
Morulas in Other Species
Mouse Morula
In the mouse, during transition from morula to blastocyst stage, differentiation of the inner cell mass (ICM) and trophectoderm (TE) has been shown to be regulated by the Hippo pathway.[4]
- Links: Mouse Development
Sea Urchin Morula
Sea Urchin early embryo cleavage pattern (SDB Gallery Images)
- Links: Sea Urchin Development
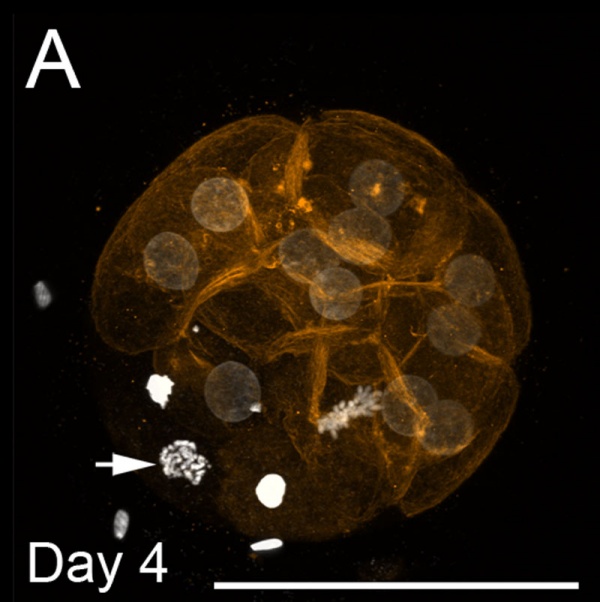
Bovine Morula
Bovine Morula[10]
- Image shows DNA staining (white) and f-actin filaments (orange) at day 4. Scale bars represent 100 µm.
- Pale staining round nuclei are at interphase.
- Arrow shows single nucleus at prophase.
- A single nucleus is seen at metaphase.
- Condensed bright nuclei are apoptotic.
- Links: Bovine Development | mitosis
Morula Biopsy
Biopsy of compact morula-stage embryos[11]
- (A) A compact morula-stage embryo before biopsy.
- (B–G) Steps of the biopsy.
- (H) An embryo at 2 h after biopsy.
References
- ↑ 1.0 1.1 Zhang P, Zucchelli M, Bruce S, Hambiliki F, Stavreus-Evers A, Levkov L, Skottman H, Kerkelä E, Kere J & Hovatta O. (2009). Transcriptome profiling of human pre-implantation development. PLoS ONE , 4, e7844. PMID: 19924284 DOI.
- ↑ Milachich T. (2014). New advances of preimplantation and prenatal genetic screening and noninvasive testing as a potential predictor of health status of babies. Biomed Res Int , 2014, 306505. PMID: 24783200 DOI.
- ↑ 3.0 3.1 Hirate Y, Hirahara S, Inoue K, Kiyonari H, Niwa H & Sasaki H. (2015). Par-aPKC-dependent and -independent mechanisms cooperatively control cell polarity, Hippo signaling, and cell positioning in 16-cell stage mouse embryos. Dev. Growth Differ. , 57, 544-56. PMID: 26450797 DOI.
- ↑ 4.0 4.1 Emura N, Saito Y, Miura R & Sawai K. (2020). Effect of Downregulating the Hippo Pathway Members YAP1 and LATS2 Transcripts on Early Development and Gene Expression Involved in Differentiation in Porcine Embryos. Cell Reprogram , , . PMID: 32150685 DOI.
- ↑ Dunwell TL & Holland PW. (2016). Diversity of human and mouse homeobox gene expression in development and adult tissues. BMC Dev. Biol. , 16, 40. PMID: 27809766 DOI.
- ↑ 6.0 6.1 Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM & Reijo Pera RA. (2010). Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat. Biotechnol. , 28, 1115-21. PMID: 20890283 DOI.
- ↑ Galán A, Montaner D, Póo ME, Valbuena D, Ruiz V, Aguilar C, Dopazo J & Simón C. (2010). Functional genomics of 5- to 8-cell stage human embryos by blastomere single-cell cDNA analysis. PLoS ONE , 5, e13615. PMID: 21049019 DOI.
- ↑ 8.0 8.1 Dard N, Louvet-Vallée S & Maro B. (2009). Orientation of mitotic spindles during the 8- to 16-cell stage transition in mouse embryos. PLoS ONE , 4, e8171. PMID: 19997595 DOI.
- ↑ Fenwick J, Platteau P, Murdoch AP & Herbert M. (2002). Time from insemination to first cleavage predicts developmental competence of human preimplantation embryos in vitro. Hum. Reprod. , 17, 407-12. PMID: 11821286
- ↑ Leidenfrost S, Boelhauve M, Reichenbach M, Güngör T, Reichenbach HD, Sinowatz F, Wolf E & Habermann FA. (2011). Cell arrest and cell death in mammalian preimplantation development: lessons from the bovine model. PLoS ONE , 6, e22121. PMID: 21811561 DOI.
- ↑ Zakharova EE, Zaletova VV & Krivokharchenko AS. (2014). Biopsy of human morula-stage embryos: outcome of 215 IVF/ICSI cycles with PGS. PLoS ONE , 9, e106433. PMID: 25191937 DOI.
Reviewss
Articles
Bessonnard S, Mesnard D & Constam DB. (2015). PC7 and the related proteases Furin and Pace4 regulate E-cadherin function during blastocyst formation. J. Cell Biol. , 210, 1185-97. PMID: 26416966 DOI.
Dzamba BJ, Jakab KR, Marsden M, Schwartz MA & DeSimone DW. (2009). Cadherin adhesion, tissue tension, and noncanonical Wnt signaling regulate fibronectin matrix organization. Dev. Cell , 16, 421-32. PMID: 19289087 DOI.
Santos J, Pereira CF, Di-Gregorio A, Spruce T, Alder O, Rodriguez T, Azuara V, Merkenschlager M & Fisher AG. (2010). Differences in the epigenetic and reprogramming properties of pluripotent and extra-embryonic stem cells implicate chromatin remodelling as an important early event in the developing mouse embryo. Epigenetics Chromatin , 3, 1. PMID: 20157423 DOI.
Search PubMed
Search Pubmed: morula development | blastomere development |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 16) Embryology Morula Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Morula_Development
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G