Molecular Development - X Inactivation: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| Line 12: | Line 12: | ||
A second form of X inactivation that occurs only in male meiotic spermatogenesis, meiotic sex chromosome inactivation (MSCI), is not covered in these current notes. MSCI is the process of transcriptional silencing of the X and Y chromosomes.<ref name="PMID17329371"><pubmed>17329371</pubmed></ref> | A second form of X inactivation that occurs only in male meiotic spermatogenesis, meiotic sex chromosome inactivation (MSCI), is not covered in these current notes. MSCI is the process of transcriptional silencing of the X and Y chromosomes.<ref name="PMID17329371"><pubmed>17329371</pubmed></ref> | ||
:'''Links:''' [[Primordial Germ Cell Development]] | [[Genital - Female Development]] | [[:File:Human_idiogram-chromosome_X.jpg|Chromosome X]] | [[2011_Group_Project_5|2011 Group Project - Fragile X Syndrome] | |||
:'''Links:''' [[Primordial Germ Cell Development]] | [[Genital - Female Development]] | [[:File:Human_idiogram-chromosome_X.jpg|Chromosome X]] | [[2011_Group_Project_5|2011 Group Project - Fragile X Syndrome]] | |||
== Some Recent Findings == | == Some Recent Findings == | ||
| Line 68: | Line 69: | ||
The breakthrough was the discovery of the X inactive specific transcript (XIST). <ref><pubmed>1985261</pubmed></ref> | The breakthrough was the discovery of the X inactive specific transcript (XIST). <ref><pubmed>1985261</pubmed></ref> | ||
This gene is located within the "X inactivation centre" and only expressed by the inactive X chromosome. | This gene is located within the "X inactivation centre" and only expressed by the inactive X chromosome. | ||
Unlike other genes that encode protein XIST contained no "open reading frames" (ie no codons to encode amino acids). | |||
XIST is transcribed but not translated. | XIST is transcribed but not translated. | ||
XIST appears to act as RNA. Current thinking is that it binds to the X Chromosome and is involved in it's translocation to the nuclear periphery. | XIST appears to act as RNA. Current thinking is that it binds to the X Chromosome and is involved in it's translocation to the nuclear periphery. | ||
It now appears that XIST appears to initiate X inactivation and it is the methylation of the inactive X genes that maintains inactivity. | It now appears that XIST appears to initiate X inactivation and it is the methylation of the inactive X genes that maintains inactivity. | ||
| Line 80: | Line 83: | ||
DXPas34 is made up of 34bp repeats containing Ctcf motifs and is located within a CpG-rich region in the 5-prime end of the mouse Tsix gene. This region also has bidirectional promoter activity, produced overlapping forward and reverse transcripts, and is necessary for both random and imprinted X inactivation in mice. The human sequence also has a 14 kb insertion not found in mouse Tsix. | DXPas34 is made up of 34bp repeats containing Ctcf motifs and is located within a CpG-rich region in the 5-prime end of the mouse Tsix gene. This region also has bidirectional promoter activity, produced overlapping forward and reverse transcripts, and is necessary for both random and imprinted X inactivation in mice. The human sequence also has a 14 kb insertion not found in mouse Tsix. | ||
Revision as of 11:40, 1 June 2013
Introduction
The presence in females of 2 X chromosome raises the issue of gene dosage, in the case of mammals this is regulated by inactivating one of the X chromosomes. To balance expression with the autosomal chromosomes the dosage imbalance is then adjusted by doubling expression of X-linked genes in both sexes.
In some other species compensation occurs by increasing the expression of X in males. The pattern of which X chromosome is inactivated in cells appears to be random, generating 50% cells expressing Father X, 50% cells expressing Mother X (mosaic pattern). The theory of random X inactivation was first suggested in mice in 1961.[3] (More? Mary Lyon)
The process of inactivation relies on the Xist RNA, a 17 kb non-coding RNA, which accumulates on the future inactive X chromosome.
A second form of X inactivation that occurs only in male meiotic spermatogenesis, meiotic sex chromosome inactivation (MSCI), is not covered in these current notes. MSCI is the process of transcriptional silencing of the X and Y chromosomes.[4]
- Links: Primordial Germ Cell Development | Genital - Female Development | Chromosome X | 2011 Group Project - Fragile X Syndrome
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Xist <pubmed limit=5>Xist</pubmed> Search term: X inactivation <pubmed limit=5>X inactivation</pubmed> |
X inactivation Xist
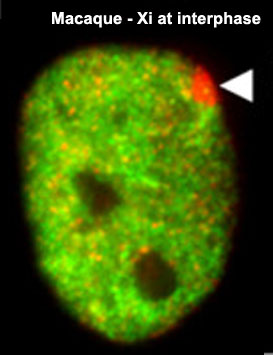
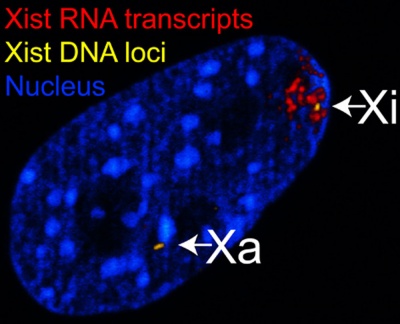
The physical region at the nucleus periphery where the inactive X chromosome is located in a female cell is described as the Barr body.[9]
|
|
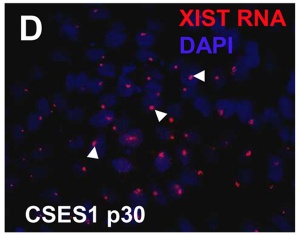
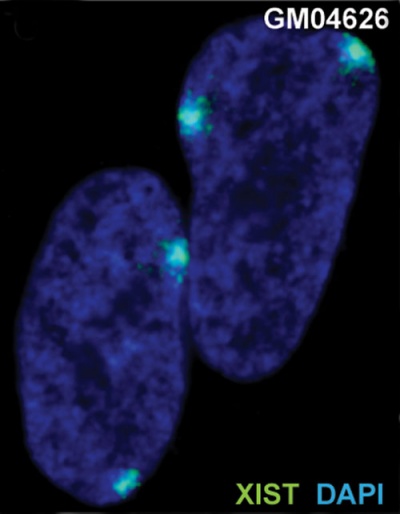
| The above figure shows confocal images from a combined RNA-DNA FISH experiment for Xist in female mouse fibroblast cells (Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse.[10] | XIST expression in two cell nuclei using RNA FISH probes for the human XIST RNA (green) in the human female fibroblast line GM04626 (karyotype: 47,XXX).[11] As there are 3 copies of the X chromosome in each cell, there are 2 inactive X chromosomes as shown by Xist staining. |
- Links: OMIM - Xist
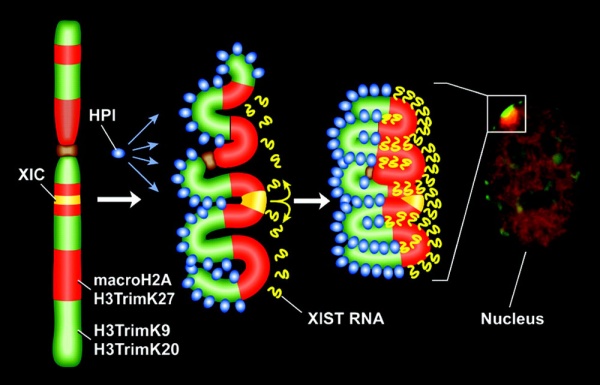
X Inactivation Model
Model for XIST RNA spread from X inactivation center[12]
Above model showing how heterochromatin of the Xi could transition between metaphase and interphase to be organized into the two nonoverlapping heterochromatin territories and to explain how XIST RNA could rapidly spread in cis outward from the X inactivation center (XIC) along only part of the Xi.
X Inactivation (xist) History
The theory of random X inactivation was first suggested in mice in 1961.[3] The breakthrough was the discovery of the X inactive specific transcript (XIST). [13] This gene is located within the "X inactivation centre" and only expressed by the inactive X chromosome. Unlike other genes that encode protein XIST contained no "open reading frames" (ie no codons to encode amino acids).
XIST is transcribed but not translated. XIST appears to act as RNA. Current thinking is that it binds to the X Chromosome and is involved in it's translocation to the nuclear periphery. It now appears that XIST appears to initiate X inactivation and it is the methylation of the inactive X genes that maintains inactivity.
- Links: Mary Lyon - X Inactivation
Tsix
A gene antisense to Xist, hence the name which is Xist backwards, in embryonic stem cells Xist repression occurs and Tsix is upregulated. This is part of the overall process of maintaining pluripotency in these cells. Tsix upregulation depends on the recruitment of another pluripotent marker Rex1, and of the associated factors Klf4 and c-Myc, by the DXPas34 minisatellite associated with the Tsix promoter.[14]
DXPas34 is made up of 34bp repeats containing Ctcf motifs and is located within a CpG-rich region in the 5-prime end of the mouse Tsix gene. This region also has bidirectional promoter activity, produced overlapping forward and reverse transcripts, and is necessary for both random and imprinted X inactivation in mice. The human sequence also has a 14 kb insertion not found in mouse Tsix.
- Links: OMIM - Tsix
Meiotic Sex Chromosome Inactivation
This second form of X inactivation takes place in the male, during spermatogenesis, as germ cells enter meiosis. This is thought to be a form of meiotic silencing of unsynapsed chromatin that silences chromosomes that fail to pair with their homologous partners.[15]
- Links: Meiosis
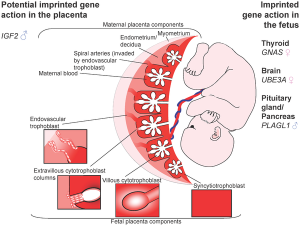
Placenta
In the mouse placenta, X inactivation is imprinted and the paternal X chromosome is always inactive. In the human placenta, initial studies appear to show a pattern of random X inactivation.[16] They also identified a preferential expression of maternal alleles, indicating that, although imprinted X chromosome inactivation has been lost during evolution, a proliferative advantage may remain for cells that inactivate the paternal X (Xp) in human placenta.
- Links: Placenta Development | Mouse Development
References
- ↑ 1.0 1.1 <pubmed>20593031</pubmed>
- ↑ McLaughlin CR, Chadwick BP. Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biol. 2011 Apr 13;12(4):R37. PMID: 21489251 | Genome Biol.
- ↑ 3.0 3.1 <pubmed>13764598</pubmed>
- ↑ <pubmed>17329371</pubmed>
- ↑ <pubmed>22069183</pubmed>
- ↑ <pubmed>21070966</pubmed>
- ↑ <pubmed>20532033</pubmed>
- ↑ <pubmed>19887628</pubmed>
- ↑ <pubmed>13865187</pubmed>
- ↑ <pubmed>21047393</pubmed>| BMC Genomics
- ↑ <pubmed>20520737</pubmed>| PLoS One
- ↑ <pubmed>15574503</pubmed>| PNAS
- ↑ <pubmed>1985261</pubmed>
- ↑ <pubmed>21085182</pubmed>
- ↑ <pubmed>17329371</pubmed>
- ↑ <pubmed>20532033</pubmed>| PLoS One.
Reviews
<pubmed>20573260</pubmed> <pubmed>19815084</pubmed> <pubmed>19802705</pubmed>
Articles
<pubmed>21811421</pubmed> <pubmed>20593031</pubmed>
Search Pubmed
July 2010 "X Inactivation" - All (3157) Review (519) Free Full Text (1066)
Search Pubmed Now: X Inactivation | Xist | Tsix | Meiotic Sex Chromosome Inactivation
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 16) Embryology Molecular Development - X Inactivation. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Molecular_Development_-_X_Inactivation
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G