File:DNA targeting platforms genome editing.jpg
Original file (922 × 800 pixels, file size: 141 KB, MIME type: image/jpeg)
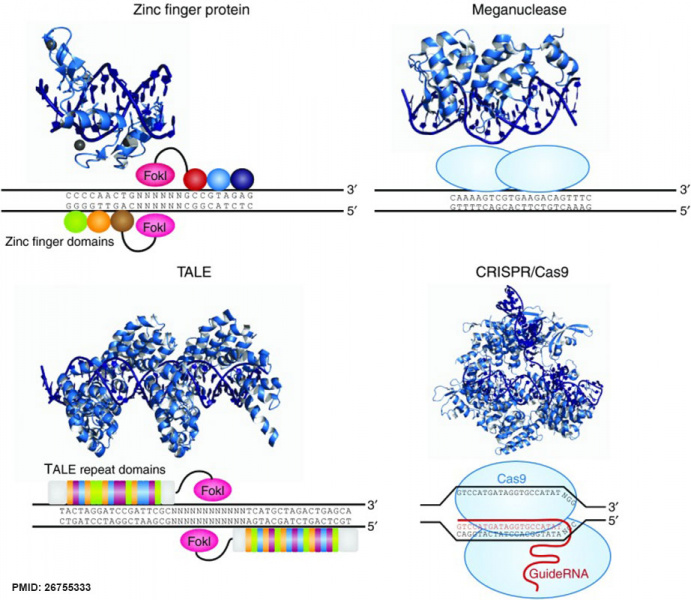
Common DNA Targeting Platforms for Genome Editing
Zinc finger nucleases
Zinc finger (ZF) proteins are the most abundant class of transcription factors and the Cys2-His2 zinc finger domain is one of the most common DNA-binding domains encoded in the human genome. The crystal structure of Zif268 has served as the basis for understanding DNA recognition by zinc fingers. In the presence of a zinc atom, the zinc finger domain forms a compact ββα structure with the α-helical portion of each finger making contact with 3 or 4 bp in the major groove of the DNA. Tandem fingers in a zinc finger array wrap around the DNA to bind extended target sequences such that a three-finger protein binds a 9 bp target site.
TALENs
The discovery of a simple one-to-one code dictating the DNA-binding specificity of TALE proteins from the plant pathogen Xanthomonas again raised the exciting possibility for modular design of novel DNA-binding proteins.114,115 Highly conserved 33–35 amino acid TALE repeats each bind a single base pair of DNA with specificity dictated by two hypervariable residues. Crystal structures of TALEs bound to DNA revealed that each repeat forms a two-helix structure connected by a loop which presents the hypervariable residue into the major groove as the protein wraps around the DNA in a superhelical structure. These modular TALE repeats can be linked together to build long arrays with custom DNA-binding specificities.
Meganucleases
Meganuclease technology involves re-engineering the DNA-binding specificity of naturally occurring homing endonucleases. The largest class of homing endonucleases is the LAGLIDADG family, which includes the well-characterized and commonly used I-CreI and I-SceI enzymes.140 Through a combination of rational design and selection, these homing endonucleases can be re-engineered to target novel sequences.
CRISPR/Cas nucleases
CRISPR-Cas RNA-guided nucleases are derived from an adaptive immune system that evolved in bacteria to defend against invading plasmids and viruses. Decades of work investigating CRISPR systems in various microbial species has elucidated a mechanism by which short sequences of invading nucleic acids are incorporated into CRISPR loci. They are then transcribed and processed into CRISPR RNAs (crRNAs) which, together with a trans-activating crRNAs (tracrRNAs), complex with CRISPR-associated (Cas) proteins to dictate specificity of DNA cleavage by Cas nucleases through Watson-Crick base pairing between nucleic acids. Building off of two studies showing that the three components required for the type II CRISPR nuclease system are the Cas9 protein, the mature crRNA and the tracrRNA, Doudna, Charpentier and colleagues showed through in vitro DNA cleavage experiments that this system could be reduced to two components by fusion of the crRNA and tracrRNA into a single guide RNA (gRNA). Furthermore, they showed that re-targeting of the Cas9/gRNA complex to new sites could be accomplished by altering the sequence of a short portion of the gRNA. Thereafter, a series of publications demonstrated that the CRISPR/Cas9 system could be engineered for efficient genetic modification in mammalian cells. Collectively these studies have propelled the CRISPR/Cas9 technology into the spotlight of the genome-editing field.
(text extract above from original article)
Reference
<pubmed>26755333 </pubmed>
Copyright
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Figure 2
Cite this page: Hill, M.A. (2024, April 18) Embryology DNA targeting platforms genome editing.jpg. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/File:DNA_targeting_platforms_genome_editing.jpg
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 11:06, 28 July 2017 |  | 922 × 800 (141 KB) | Z8600021 (talk | contribs) | ==Common DNA Targeting Platforms for Genome Editing== '''Zinc finger nucleases''' Zinc finger (ZF) proteins are the most abundant class of transcription factors and the Cys2-His2 zinc finger domain is one of the most common DNA-binding domains encod... |
You cannot overwrite this file.
File usage
The following page uses this file: