File:Bovine blastocyst 01.jpg: Difference between revisions
(uploaded a new version of "File:Bovine blastocyst 01.jpg": resized image) |
No edit summary |
||
| Line 21: | Line 21: | ||
[[Category:Bovine | [[Category:Bovine]] [[Category:Blastocyst]] | ||
Revision as of 13:26, 4 November 2011
Bovine Blastocyst
G: Cell death in the inner cell mass of bovine IVF blastocysts examined at day 7: In expanding and hatching blastocysts, DAPI staining reveals highly condensed chromatin structures that are irregularly shaped and variable in size referring to different modes and stages of cell death (arrows).
Image is a maximum intensity z-projection of a 20 µm image stack
The embryos were fixed and mounted on coverslips in such a way that the three-dimensional structure was maintained. DNA staining with DAPI is shown in white, f-actin filaments (phalloidin-TRITC) in orange.
Scale bars represent 100 µm (overviews) or 10 µm (details).
- Links: Bovine Development
Figure 2. CLSM analysis (Panel G cropped from full image)
Reference
<pubmed>21811561</pubmed>| PLoS One.
Copyright: © 2011 Leidenfrost et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 13:23, 4 November 2011 | 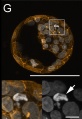 | 688 × 1,000 (99 KB) | S8600021 (talk | contribs) | resized image |
| 13:22, 4 November 2011 | 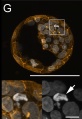 | 516 × 750 (65 KB) | S8600021 (talk | contribs) | ==Bovine Blastocyst== G: Cell death in the inner cell mass of bovine IVF blastocysts examined at day 7: In expanding and hatching blastocysts, DAPI staining reveals highly condensed chromatin structures that are irregularly shaped and variable in size re |
You cannot overwrite this file.
File usage
The following 2 pages use this file: