Introduction

Notch structure cartoon
[1]The notch proteins were first identified in drosophila development and have since been identified as regulators of cell fate decisions during development. These are a family of cell surface transmembrane receptors that pass once through the plasma membrane.
- Notch Links: Notch structure cartoon | Notch signaling pathway cartoon | Notch and signaling pathway cartoon | Developmental Signals - Notch | Molecular Factors
Some Recent Findings
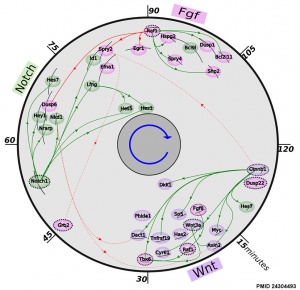
Mouse somitogenesis genes
[2]
- Notch-mediated lateral inhibition regulates proneural wave propagation when combined with EGF-mediated reaction diffusion[3] "Notch-mediated lateral inhibition regulates binary cell fate choice, resulting in salt and pepper patterns during various developmental processes. However, how Notch signaling behaves in combination with other signaling systems remains elusive. The wave of differentiation in the Drosophila visual center or "proneural wave" accompanies Notch activity that is propagated without the formation of a salt and pepper pattern, implying that Notch does not form a feedback loop of lateral inhibition during this process. However, mathematical modeling and genetic analysis clearly showed that Notch-mediated lateral inhibition is implemented within the proneural wave. Because partial reduction in EGF signaling causes the formation of the salt and pepper pattern, it is most likely that EGF diffusion cancels salt and pepper pattern formation in silico and in vivo. Moreover, the combination of Notch-mediated lateral inhibition and EGF-mediated reaction diffusion enables a function of Notch signaling that regulates propagation of the wave of differentiation."
- Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors[4] "Basal cells are multipotent airway progenitors that generate distinct epithelial cell phenotypes crucial for homeostasis and repair of the conducting airways. Little is known about how these progenitor cells expand and transition to differentiation to form the pseudostratified airway epithelium in the developing and adult lung. Here, we show by genetic and pharmacological approaches that endogenous activation of Notch3 signaling selectively controls the pool of undifferentiated progenitors of upper airways available for differentiation. This mechanism depends on the availability of Jag1 and Jag2, and is key to generating a population of parabasal cells that later activates Notch1 and Notch2 for secretory-multiciliated cell fate selection." Respiratory System Development
- Notch regulation of myogenic versus endothelial fates of cells that migrate from the somite to the limb[5] "Multipotent Pax3-positive (Pax3(+)) cells in the somites give rise to skeletal muscle and to cells of the vasculature. We had previously proposed that this cell-fate choice depends on the equilibrium between Pax3 and Foxc2 expression. In this study, we report that the Notch pathway promotes vascular versus skeletal muscle cell fates. ...We now demonstrate that in addition to the inhibitory role of Notch signaling on skeletal muscle cell differentiation, the Notch pathway affects the Pax3:Foxc2 balance and promotes the endothelial versus myogenic cell fate, before migration to the limb, in multipotent Pax3(+) cells in the somite of the mouse embryo." Limb Development | Muscle Development
- The precise timeline of transcriptional regulation reveals causation in mouse somitogenesis network[2] "In vertebrate development, the segmental pattern of the body axis is established as somites, masses of mesoderm distributed along the two sides of the neural tube, are formed sequentially in the anterior-posterior axis. This mechanism depends on waves of gene expression associated with the Notch, Fgf and Wnt pathways."
- Role of p63 and the Notch pathway in cochlea development and sensorineural deafness[6] "The ectodermal dysplasias are a group of inherited autosomal dominant syndromes associated with heterozygous mutations in the Tumor Protein p63 (TRP63) gene. Here we show that, in addition to their epidermal pathology, a proportion of these patients have distinct levels of deafness. ...these data demonstrate that TAp63, acting via the Notch pathway, is crucial for the development of the organ of Corti, providing a molecular explanation for the sensorineural deafness in ectodermal dysplasia patients with TRP63 mutations." Sensory - Hearing and Balance Development
- Notch signaling regulates late-stage epidermal differentiation and maintains postnatal hair cycle homeostasis [7] "Notch signaling involves ligand-receptor interactions through direct cell-cell contact. Multiple Notch receptors and ligands are expressed in the epidermis and hair follicles during embryonic development and the adult stage. Although Notch signaling plays an important role in regulating differentiation of the epidermis and hair follicles, it remains unclear how Notch signaling participates in late-stage epidermal differentiation and postnatal hair cycle homeostasis. ...our data reveal a role for Notch signaling in regulating late-stage epidermal differentiation. Notch signaling is required for postnatal hair cycle homeostasis by maintaining proper proliferation and differentiation of hair follicle stem cells."
- Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development [8] "The role of Notch signaling in cartilage differentiation and maturation in vivo was examined. Conditional Notch pathway gain and loss of function was achieved using a Cre/loxP approach to manipulate Notch signaling in cartilage precursors and chondrocytes of the developing mouse embryo. Conditional overexpression of activated Notch intracellular domain (NICD) in the chondrocyte lineage results in skeletal malformations with decreased cartilage precursor proliferation and inhibited hypertrophic chondrocyte differentiation. Likewise, expression of NICD in cartilage precursors inhibits sclerotome differentiation, resulting in severe axial skeleton abnormalities. Furthermore, conditional loss of Notch signaling via RBP-J gene deletion in the chondrocyte lineage results in increased chondrocyte proliferation and skeletal malformations consistent with the observed increase in hypertrophic chondrocytes. In addition, the Notch pathway inhibits expression of Sox9 and its target genes required for normal chondrogenic cell proliferation and differentiation. Together, our results demonstrate that appropriate Notch pathway signaling is essential for proper chondrocyte progenitor proliferation and for the normal progression of hypertrophic chondrocyte differentiation into bone in the developing appendicular and axial skeletal elements." Cartilage Development
- Notch signalling in the paraxial mesoderm is most sensitive to reduced Pofut1 levels during early mouse development[9] "Notch-dependent processes apparently differ with respect to their requirement for levels of POFUT1. Normal Lfng expression and anterior-posterior somite patterning is highly sensitive to reduced POFUT1 levels in early mammalian embryos, whereas other early Notch-dependent processes such as establishment of left-right asymmetry or neurogenesis are not. Thus, it appears that in the presomitic mesoderm (PSM) Notch signalling is particularly sensitive to POFUT1 levels. Reduced POFUT1 levels might affect Notch trafficking or overall O-fucosylation. Alternatively, reduced O-fucosylation might preferentially affect sites that are substrates for LFNG and thus important for somite formation and patterning."
|
| More recent papers
|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
- This search now requires a manual link as the original PubMed extension has been disabled.
- The displayed list of references do not reflect any editorial selection of material based on content or relevance.
- References also appear on this list based upon the date of the actual page viewing.
References listed on the rest of the content page and the associated discussion page (listed under the publication year sub-headings) do include some editorial selection based upon both relevance and availability.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References
Search term: Notch
<pubmed limit=5>Notch</pubmed>
|
Notch Signaling
|
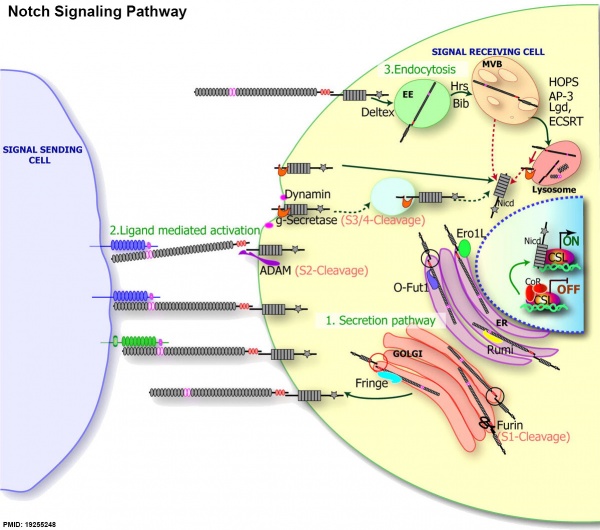
Notch signaling pathway[1]
- Secretory pathway: the modifications during the secretion of Notch to the membrane in the ER (purple) and the Golgi (orange). Notch is translated inside ER, where it is glycosylated by an O-fucosyltransferase O-Fut1 (light purple) and O-glucosyltransferase Rumi (yellow). Note the black circle on the Notch molecule in the ER; because Notch is not cleaved, the extracellular and intracellular domains are physically linked. Notch is then translocated into Golgi, where it is cleaved by Furin protease (scissors) at the S1 site and further modified by the N-acetylglucosaminyltransferase, Fringe. Note the red circle on the Notch molecule in the Golgi after S1 cleavage; the extracellular and intracellular domains are not physically linked
- Ligand-mediated activation: Notch (gray) interacts with the DSL ligands, Delta (blue) and Serrate (green), resulting in a series of proteolytic cleavage events induced by ligand binding. The S2 cleavage is mediated by ADAM protease (purple), whereas the S3/4 cleavage event is mediated by γ-secretase (g-secretase, in orange). Several studies also suggest that γ-secretase–mediated cleavage can occur inside endocytic compartment (shown in the light blue circle).
- The endocytic regulation of the Notch receptor: full-length Notch can undergo endocytosis, leading to translocation of Notch into EEs, MVB, and lysosomes. From genetic data, several proteins have been identified to modulate this process, including Hrs and Bib, possibly during the EE-to-MVB transition of Notch, Lgd, and ESCRT complex, or during the MVB-to-lysosomes transition. These proteins further modulate Notch activity as described in the text. The dotted red arrow shows that in mutants that affect trafficking from the MVB to the lysosome, or if Notch is not trafficked to the lumen of the lysosome, Notch can undergo γ-secretase cleavage, resulting in a Notch GOF phenotypes.
(text from original figure legend)
|
Notch Receptors
NOTCH1
NOTCH2
NOTCH3
- Notch3 activation retains mammary luminal cell in a nonproliferative state.[10]
NOTCH4
Notch Ligands
Functions
Developmental patterning signal.
Spinal Cord Development
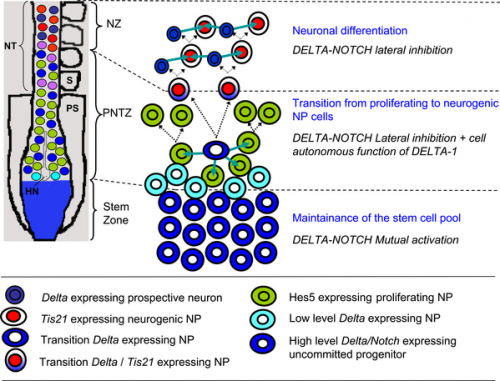
Model of the embryonic rostro-caudal gradient of neurogenesis along the chicken spinal cord from the stem zone to the neurogenic neural tube summarising how DELTA-NOTCH signalling may be involved in these processes.[11]
- Caudal to rostral decreasing FGF gradient, leads to Delta-1 expression decrease in cells that leave the stem zone (light blue) and move into the PNTZ where they intermingle with cells that do not express Delta-1.
- Generates differences in DELTA/NOTCH signalling between adjacent cells that may initiate lateral inhibition.
- Upregulation of Delta-1 in single NP cells which signal (blue arrows) and activate NOTCH signalling in adjacent cells, which as a consequence express Hes5 and are maintained in a proliferating state.
- Delta-1 expressing NP cell divides into two cells that express Tis21.
- Double Delta-1/Tis21 labelled NP down regulate the expression of Delta-1 as they reach the NZ and begin to divide in a neurogenic manner.
- One of the daughter cells upregulates Delta-1 expression and differentiates as a neuron while the other one, which receives NOTCH signalling (blue arrows), remains as neurogenic NP. Hensen node (HN), neural tube (NT), neurogenic zone (NZ), proliferation to neurogenesis transition zone (PNTZ), presomitic territory (PS), somite (S).
|
|
Endoderm Development
Endoderm differentiates to form the respiratory airway epithelium and glands. This epithelium is continuously replaced through life from a basal cell pool of undifferentiated airway progenitors. A recent study[4] has shown that the progenitor pool is regulated by the Notch3-Jagged signaling pathway. The mechanism appears dependent upon the availability of Jag1 and Jag2 (generating parabasal cells) that later activates Notch1 and Notch2 leading to a secretory-multiciliated cell fate.
- L;inks: Respiratory System Development
Mesoderm Development
Cartilage Development
Muscle Regeneration
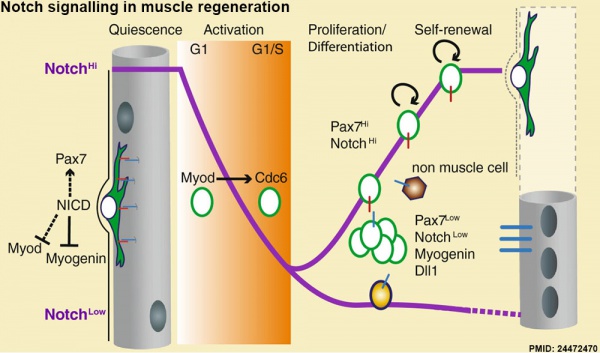
Notch signalling in muscle regeneration[12]
Hypothalamus Development
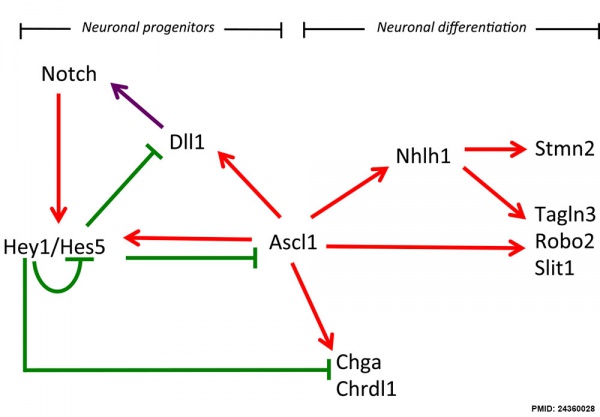
Hypothalamus Development Gene Interaction Model[13]
- Links: Hypothalamus Development
Abnormalities
Alagille Syndrome
Mutations in the human homolog of Jagged-1 (JAG1) located on chromosome 20p12 cause Alagille Syndrome. Abnormalities are seen in gastrointestinal (liver cholestasis), cardiac (heart), renal (kidney), skeletal, ocular, and facial systems.
- Links: Alagille Syndrome
References
- ↑ 1.0 1.1 <pubmed>19255248</pubmed>
- ↑ 2.0 2.1 <pubmed>24304493</pubmed>| BMC Dev Biol.
- ↑ <pubmed>27535937</pubmed>
- ↑ 4.0 4.1 <pubmed>25564622</pubmed>
- ↑ <pubmed>24927569</pubmed>
- ↑ <pubmed>20513039</pubmed>
- ↑ <pubmed>19590010</pubmed>
- ↑ <pubmed>19590010</pubmed>
- ↑ <pubmed>19161597</pubmed>
- ↑ <pubmed>24100291</pubmed>
- ↑ <pubmed>18000541</pubmed>| PLoS ONE
- ↑ <pubmed>24472470</pubmed>| BMC Dev Biol.
- ↑ 24360028<pubmed>24360028</pubmed>| Neural Dev.
Reviews
<pubmed></pubmed>
<pubmed></pubmed>
<pubmed></pubmed>
<pubmed>23165243</pubmed>
<pubmed> 22399351</pubmed>
<pubmed>22397947</pubmed>
<pubmed>21828089</pubmed>
Search Pubmed
Search Bookshelf Notch
Search Pubmed Now: Notch Signaling
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 18) Embryology Developmental Signals - Notch. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Developmental_Signals_-_Notch
- What Links Here?
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G






