|
|
| Line 54: |
Line 54: |
|
| |
|
| Notch signalling in muscle regeneration<ref><pubmed>24472470</pubmed>| [http://www.biomedcentral.com/1471-213X/14/2 BMC Dev Biol.]</ref> | | Notch signalling in muscle regeneration<ref><pubmed>24472470</pubmed>| [http://www.biomedcentral.com/1471-213X/14/2 BMC Dev Biol.]</ref> |
| | |
| | ===Hypothalamus Development=== |
| | |
| | [[File:Hypothalamus gene interaction model.jpg|600px]] |
| | |
| | Hypothalamus Development Gene Interaction Model<ref name=PMID>24360028<pubmed>24360028</pubmed>| [http://www.neuraldevelopment.com/content/8//25 Neural Dev.]</ref> |
|
| |
|
| ==References== | | ==References== |
Revision as of 22:54, 23 March 2014
Introduction
The notch proteins were first identified in drosophila development and have since been identified as regulators of cell fate decisions during development. These are a family of cell surface transmembrane receptors that pass once through the plasma membrane.
Some Recent Findings
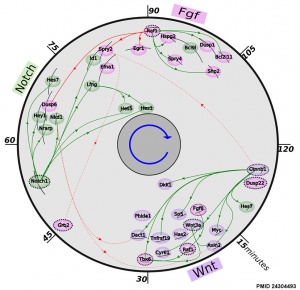
Mouse somitogenesis genes
[1]
- The precise timeline of transcriptional regulation reveals causation in mouse somitogenesis network[1] "In vertebrate development, the segmental pattern of the body axis is established as somites, masses of mesoderm distributed along the two sides of the neural tube, are formed sequentially in the anterior-posterior axis. This mechanism depends on waves of gene expression associated with the Notch, Fgf and Wnt pathways."
- Role of p63 and the Notch pathway in cochlea development and sensorineural deafness[2] "The ectodermal dysplasias are a group of inherited autosomal dominant syndromes associated with heterozygous mutations in the Tumor Protein p63 (TRP63) gene. Here we show that, in addition to their epidermal pathology, a proportion of these patients have distinct levels of deafness. ...these data demonstrate that TAp63, acting via the Notch pathway, is crucial for the development of the organ of Corti, providing a molecular explanation for the sensorineural deafness in ectodermal dysplasia patients with TRP63 mutations." Sensory - Hearing and Balance Development
- Notch signaling regulates late-stage epidermal differentiation and maintains postnatal hair cycle homeostasis [3] "Notch signaling involves ligand-receptor interactions through direct cell-cell contact. Multiple Notch receptors and ligands are expressed in the epidermis and hair follicles during embryonic development and the adult stage. Although Notch signaling plays an important role in regulating differentiation of the epidermis and hair follicles, it remains unclear how Notch signaling participates in late-stage epidermal differentiation and postnatal hair cycle homeostasis. ...our data reveal a role for Notch signaling in regulating late-stage epidermal differentiation. Notch signaling is required for postnatal hair cycle homeostasis by maintaining proper proliferation and differentiation of hair follicle stem cells."
- Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development [4] "The role of Notch signaling in cartilage differentiation and maturation in vivo was examined. Conditional Notch pathway gain and loss of function was achieved using a Cre/loxP approach to manipulate Notch signaling in cartilage precursors and chondrocytes of the developing mouse embryo. Conditional overexpression of activated Notch intracellular domain (NICD) in the chondrocyte lineage results in skeletal malformations with decreased cartilage precursor proliferation and inhibited hypertrophic chondrocyte differentiation. Likewise, expression of NICD in cartilage precursors inhibits sclerotome differentiation, resulting in severe axial skeleton abnormalities. Furthermore, conditional loss of Notch signaling via RBP-J gene deletion in the chondrocyte lineage results in increased chondrocyte proliferation and skeletal malformations consistent with the observed increase in hypertrophic chondrocytes. In addition, the Notch pathway inhibits expression of Sox9 and its target genes required for normal chondrogenic cell proliferation and differentiation. Together, our results demonstrate that appropriate Notch pathway signaling is essential for proper chondrocyte progenitor proliferation and for the normal progression of hypertrophic chondrocyte differentiation into bone in the developing appendicular and axial skeletal elements."
- Notch signalling in the paraxial mesoderm is most sensitive to reduced Pofut1 levels during early mouse development[5] "Notch-dependent processes apparently differ with respect to their requirement for levels of POFUT1. Normal Lfng expression and anterior-posterior somite patterning is highly sensitive to reduced POFUT1 levels in early mammalian embryos, whereas other early Notch-dependent processes such as establishment of left-right asymmetry or neurogenesis are not. Thus, it appears that in the presomitic mesoderm (PSM) Notch signalling is particularly sensitive to POFUT1 levels. Reduced POFUT1 levels might affect Notch trafficking or overall O-fucosylation. Alternatively, reduced O-fucosylation might preferentially affect sites that are substrates for LFNG and thus important for somite formation and patterning."
|
| More recent papers
|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
- This search now requires a manual link as the original PubMed extension has been disabled.
- The displayed list of references do not reflect any editorial selection of material based on content or relevance.
- References also appear on this list based upon the date of the actual page viewing.
References listed on the rest of the content page and the associated discussion page (listed under the publication year sub-headings) do include some editorial selection based upon both relevance and availability.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References
Search term: Notch
<pubmed limit=5>Notch</pubmed>
|
Notch Receptors
NOTCH1
NOTCH2
NOTCH3
NOTCH4
Notch Ligands
Functions
Developmental patterning signal.
Mesoderm Development
Cartilage Development
Muscle Regeneration
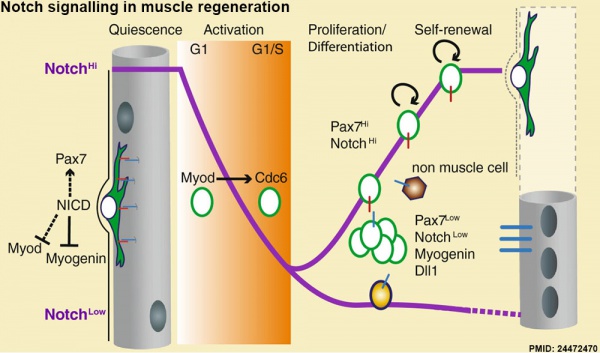
Notch signalling in muscle regeneration[6]
Hypothalamus Development
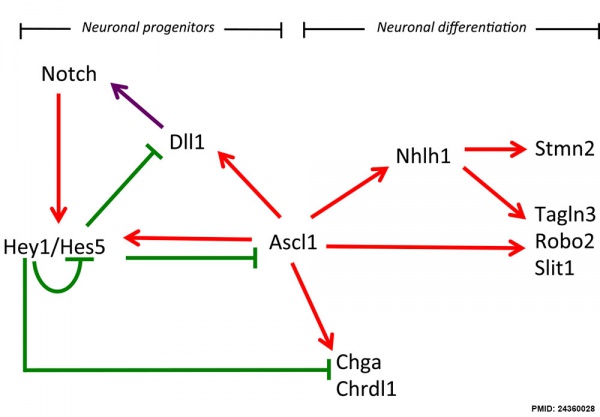
Hypothalamus Development Gene Interaction Model[7]
References
- ↑ 1.0 1.1 <pubmed>24304493</pubmed>| BMC Dev Biol.
- ↑ <pubmed>20513039</pubmed>
- ↑ <pubmed>19590010</pubmed>
- ↑ <pubmed>19590010</pubmed>
- ↑ <pubmed>19161597</pubmed>
- ↑ <pubmed>24472470</pubmed>| BMC Dev Biol.
- ↑ 24360028<pubmed>24360028</pubmed>| Neural Dev.
Reviews
<pubmed></pubmed>
<pubmed></pubmed>
<pubmed></pubmed>
<pubmed>23165243</pubmed>
<pubmed> 22399351</pubmed>
<pubmed>22397947</pubmed>
<pubmed>21828089</pubmed>
Search Pubmed
Search Bookshelf Notch
Search Pubmed Now: Notch Signaling
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 25) Embryology Developmental Signals - Notch. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Developmental_Signals_-_Notch
- What Links Here?
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G



