Developmental Signals - Hippo: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| Line 6: | Line 6: | ||
Hippo is a protein kinase cascade pathway, getting its name from the “hippopotamus”-like phenotype. | Hippo is a protein kinase cascade pathway, getting its name from the “hippopotamus”-like phenotype. | ||
{| | |||
! Fly Phenotype (dorsal view head thorax SEM) | |||
|- | |||
| [[:File:Fly Hippo-type dorsal view head thorax SEM.jpg|300px]] | |||
| [[File:Fly WT dorsal view head thorax SEM.jpg|300px]] | |||
|- | |||
| Hippo-type (hpo) | |||
| Wild-type (WT) | |||
|- | |||
| | |||
| Image source<ref name="PMID24336504"><pubmed>24336504</pubmed>| [http://www.nature.com/nrd/journal/v13/n1/full/nrd4161.html Nat Rev Drug Discov.]</ref> | |||
|} | |||
{{Factor Links}} | [[:Category:Hippo|Category:Hippo]] | {{Factor Links}} | [[:Category:Hippo|Category:Hippo]] | ||
Revision as of 11:01, 11 June 2015
| Embryology - 16 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
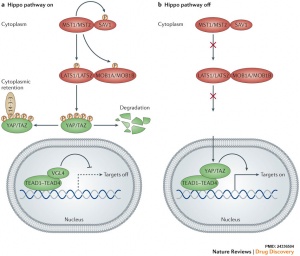
The Hippo (Hpo) pathway, first identified in Drosophila, controls organ size by regulating cell proliferation (inhibition) and apoptosis (induction). In contrast, the TOR signalling pathway regulates organ size by stimulating cell growth, thus increasing cell size.
Hippo is a protein kinase cascade pathway, getting its name from the “hippopotamus”-like phenotype.
| Fly Phenotype (dorsal view head thorax SEM) | |
|---|---|
| 300px | 
|
| Hippo-type (hpo) | Wild-type (WT) |
| Image source[1] |
| Factor Links: AMH | hCG | BMP | sonic hedgehog | bHLH | HOX | FGF | FOX | Hippo | LIM | Nanog | NGF | Nodal | Notch | PAX | retinoic acid | SIX | Slit2/Robo1 | SOX | TBX | TGF-beta | VEGF | WNT | Category:Molecular |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Embryo Hippo | Images <pubmed limit=5>Embryo Hippo</pubmed> |
Early Development
Blastocyst
| Mouse blastocyst (32 cell stage) | |
|---|---|
| phosphorylation of angiomotin at adherens junctions | cell polarization sequesters Amot from basolateral adherens junctions |
| active Hippo signaling | inactive Hippo signaling |
| inner cells | outer cells |
| inner cell mass (ICM) fate | trophectoderm (TE) fate |
- (Table data see review[2] Notch signaling also has a role in blastocyst fate development)
- Links: Blastocyst
Organ Expression
Endoderm
Fetal Gonad
Neural
References
- ↑ 1.0 1.1 <pubmed>24336504</pubmed>| Nat Rev Drug Discov.
- ↑ 2.0 2.1 <pubmed>25986053</pubmed>
Reviews
<pubmed>26032720</pubmed> <pubmed>22575479</pubmed> <pubmed>21808241</pubmed> <pubmed>19517570</pubmed>
Articles
<pubmed>25628125</pubmed> <pubmed>25918243</pubmed> <pubmed></pubmed> <pubmed></pubmed>
Search Pubmed
Search Bookshelf Hippo
Search Pubmed Now: Hippo
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 16) Embryology Developmental Signals - Hippo. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Developmental_Signals_-_Hippo
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G
