Developmental Signals - Bone Morphogenetic Protein: Difference between revisions
mNo edit summary |
|||
| (47 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Header}} | |||
==Introduction== | ==Introduction== | ||
Belongs to the transforming growth factor-beta (TGFB) superfamily, humans have [[#Human BMP Family|15 members]] in this protein signaling family. The proteins are synthesized as prepropeptides, then cleaved, and processed into dimeric proteins. | |||
:TGFB family members: TGFB1, TGFB, TGFB3, bone morphogenetic proteins Bmp-2A, Bmp-2B, Bmp-3, and Bmp-6. mullerian inhibitory substance. | |||
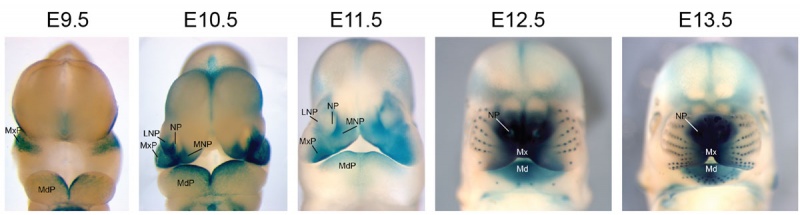
Mouse Bmp4 expression face. | [[File:Mouse Bmp4 expression face 01.jpg|800px]] | ||
Mouse Bmp4 expression face.{{#pmid:22701669|PMID22701669}} | |||
{{Mouse Bmp4}} | {{Mouse Bmp4}} | ||
Growth Differentiation Factor-6 (Gdf6) is a member of the Bone Morphogenetic Protein (BMP) family of secreted signaling molecules. | |||
{{Factor Links}} | |||
== Some Recent Findings == | == Some Recent Findings == | ||
| Line 14: | Line 21: | ||
|-bgcolor="F5FAFF" | |-bgcolor="F5FAFF" | ||
| | | | ||
* '''Developmental stalling and organ-autonomous regulation of morphogenesis''' | * '''Serum concentrations of oocyte-secreted factors BMP15 and GDF9 during IVF and in women with reproductive pathologies'''{{#pmid:31211369}} "Oocyte-secreted factors, bone morphogenetic protein-15 ({{BMP}}15) and growth differentiation factor-9 (GDF9) are critical for folliculogenesis and fertility. This study developed ELISAs for the measurement of BMP15 and GDF9 in serum and investigated their usefulness as biomarkers of female reproductive function. Serum samples were obtained from women undergoing infertility treatments (n=154), and from peri- and post-menopausal women (n=28). Serum concentrations of BMP15 and GDF9 in women relative to age, AMH, number of oocytes retrieved, and polycystic ovaries/syndrome (PCO(S)) after superovulation for IVF. BMP15 and GDF9 immunoassays were validated for specificity, sensitivity (24 and 26 pg/ml, respectively) and reproducibility. BMP15 and GDF9 were detectable in 61% and 29% of women, respectively. BMP15 and GDF9 varied 64- and 15-fold respectively between women, but did not change within subjects following ovarian stimulation with gonadotropins. Serum GDF9 concentration, but not BMP15, was associated with oocyte number retrieved in non-PCO(S) patients (p=0.018). GDF9 and BMP15 associations with oocyte number differed significantly (p<0.05) with PCO(S) status. GDF9 concentrations were lower in poor responders (women with <4 oocytes retrieved or cancelled cycles; p=0.020). Serum BMP15, but not GDF9, was lower in women over 55 years, compared to women of reproductive age (p<0.01). This study develops and validates immunoassays to quantitate BMP15 and GDF9 in human serum, and to correlate concentrations with female reproductive potential. Although assay sensitivities require improvement, this study demonstrates the diagnostic potential of oocyte-secreted BMP15 and GDF9 as serum biomarkers in reproductive medicine." | ||
* '''BMP controls dorsoventral and neural patterning in indirect-developing hemichordates providing insight into a possible origin of chordates'''{{#pmid:31189599|PMID31189599}} "A defining feature of chordates is the unique presence of a dorsal hollow neural tube that forms by internalization of the ectodermal neural plate specified via inhibition of BMP signaling during gastrulation. While BMP controls dorsoventral (DV) patterning across diverse bilaterians, the BMP-active side is ventral in chordates and dorsal in many other bilaterians. How this phylum-specific DV inversion occurs and whether it is coupled to the emergence of the dorsal neural plate are unknown. Here we explore these questions by investigating an indirect-developing enteropneust from the hemichordate phylum, which together with echinoderms form a sister group of the chordates. We found that in the hemichordate larva, BMP signaling is required for DV patterning and is sufficient to repress neurogenesis. We also found that transient overactivation of BMP signaling during gastrulation concomitantly blocked mouth formation and centralized the nervous system to the ventral ectoderm in both hemichordate and sea urchin larvae. Moreover, this mouthless, neurogenic ventral ectoderm displayed a medial-to-lateral organization similar to that of the chordate neural plate. Thus, indirect-developing deuterostomes use BMP signaling in DV and neural patterning, and an elevated BMP level during gastrulation drives pronounced morphological changes reminiscent of a DV inversion. These findings provide a mechanistic basis to support the hypothesis that an inverse chordate body plan emerged from an indirect-developing ancestor by tinkering with BMP signaling." | |||
* '''Bmp4 is an essential growth factor for the initiation of {{genita}}l tubercle (GT) outgrowth'''{{#pmid:30714224|PMID30714224}} "The external genitalia are appendage organs outgrowing from the posterior body trunk. Murine genital tubercle (GT), anlage of external genitalia, initiates its outgrowth from embryonic day (E) {{ME10.5}} as a bud structure. Several growth factors such as fibroblast growth factor (FGF), Wnt and Sonic hedgehog ({{Shh}}) are essential for the GT outgrowth. However, the mechanisms of initiation of GT outgrowth are poorly understood. We previously identified bone morphogenetic protein ({{Bmp}}) signaling as a negative regulator for GT outgrowth. We show here novel aspects of Bmp4 functions for GT outgrowth. We identified the Bmp4 was already expressed in cloaca region at E9.5, before GT outgrowth. To analyze the function of Bmp4 at early stage for the initiation of GT outgrowth, we utilized the Hoxa3-Cre driver and Bmp4 flox/flox mouse lines. Hoxa3 Cre/+ ; Bmp4 flox/flox mutant mice showed the hypoplasia of GT with reduced expression of outgrowth promoting genes such as Wnt5a, Hoxd13 and p63, whereas Shh expression was not affected. Formation of distal urethral epithelium (DUE) marked by the Fgf8 expression is essential for controlling mesenchymal genes expression in GT and subsequent its outgrowth. Furthermore, Fgf8 expression was dramatically reduced in such mutant mice indicating the defective DUE formation. Hence, current results indicate that Bmp4 is an essential growth factor for the initiation of GT outgrowth independent of Shh signaling. Thus, Bmp4 positively regulates for the formation of DUE. The current study provides new insights into the function of Bmp signaling at early stage for the initiation of GT outgrowth." {{genital}} | |||
* '''{{BMP}} and {{FGF}} signaling interact to pattern {{mesoderm}} by controlling basic helix-loop-helix transcription factor activity'''{{#pmid:29877796|PMID29877796}} "The {{mesoderm}}al germ layer is patterned into mediolateral subtypes by signaling factors including {{BMP}} and {{FGF}}. How these pathways are integrated to induce specific mediolateral cell fates is not well understood. We used mesoderm derived from post-gastrulation neuromesodermal progenitors (NMPs), which undergo a binary mediolateral patterning decision, as a simplified model to understand how FGF acts together with BMP to impart mediolateral fate. Using zebrafish and mouse NMPs, we identify an evolutionarily conserved mechanism of BMP and FGF mediated mediolateral mesodermal patterning that occurs through modulation of basic helix-loop-helix (bHLH) transcription factor activity. BMP imparts lateral fate through induction of Id helix loop helix (HLH) proteins, which antagonize bHLH transcription factors, induced by FGF signaling, that specify medial fate. We extend our analysis of zebrafish development to show that bHLH activity is responsible for the mediolateral patterning of the entire mesodermal germ layer." | |||
|} | |||
{| class="wikitable mw-collapsible mw-collapsed" | |||
! More recent papers | |||
|- | |||
| [[File:Mark_Hill.jpg|90px|left]] {{Most_Recent_Refs}} | |||
Search term: [http://www.ncbi.nlm.nih.gov/pubmed/?term=Bone+Morphogenetic+Protein ''Bone Morphogenetic Protein''] | [http://www.ncbi.nlm.nih.gov/pubmed/?term=BMP ''BMP''] | [http://www.ncbi.nlm.nih.gov/pubmed/?term=BMP4 ''BMP4''] | [http://www.ncbi.nlm.nih.gov/pubmed/?term=BMP7 ''BMP7''] | [http://www.ncbi.nlm.nih.gov/pubmed/?term=BMP15 ''BMP15''] | |||
|} | |||
{| class="wikitable mw-collapsible mw-collapsed" | |||
! Older papers | |||
|- | |||
| {{Older papers}} | |||
* '''Bone morphogenetic protein 4 promotes craniofacial neural crest induction from human pluripotent stem cells'''{{#pmid:26934293|PMID26934293}} "Neural crest (NC) cells are a group of cells located in the neural folds at the boundary between the neural and epidermal ectoderm. Cranial NC cells migrate to the branchial arches and give rise to the majority of the craniofacial region, whereas trunk and tail NC cells contribute to the heart, enteric ganglia of the gut, melanocytes, sympathetic ganglia, and adrenal chromaffin cells. ...These BMP4-treated NC cells were capable of differentiation into osteocytes and chondrocytes. The results of the present study indicate that BMP4 regulates cranial positioning during NC development." {{Neural crest}} | |||
* '''Review - Embryo Development. Bmp Gradients'''{{#pmid:26113727|PMID26113727}} "Bone morphogenetic proteins (BMPs) act in dose-dependent fashion to regulate cell fate choices in a myriad of developmental contexts. In early vertebrate and invertebrate embryos, BMPs and their antagonists establish epidermal versus central nervous system domains. In this highly conserved system, BMP antagonists mediate the neural-inductive activities proposed by Hans Spemann and Hilde Mangold nearly a century ago. BMPs distributed in gradients subsequently function as morphogens to subdivide the three germ layers into distinct territories and act to organize body axes, regulate growth, maintain stem cell niches, or signal inductively across germ layers. In this Review, we summarize the variety of mechanisms that contribute to generating reliable developmental responses to BMP gradients and other morphogen systems." | |||
* '''Construction of a vertebrate embryo from two opposing morphogen gradients'''{{#pmid:24700857|PMID24700857}} "Here, we show that opposing gradients of bone morphogenetic protein (BMP) and Nodal, two transforming growth factor family members that act as morphogens, are sufficient to induce molecular and cellular mechanisms required to organize, in vivo or in vitro, uncommitted cells of the zebrafish blastula animal pole into a well-developed embryo." {{Zebrafish}} | |||
* '''Developmental stalling and organ-autonomous regulation of morphogenesis'''{{#pmid:22084104|PMID22084104}} "Timing of organ development during embryogenesis is coordinated such that at birth, organ and fetal size and maturity are appropriately proportioned. The extent to which local developmental timers are integrated with each other and with the signaling interactions that regulate morphogenesis to achieve this end is not understood. Using the absolute requirement for a signaling pathway activity (bone morphogenetic protein, BMP) during a critical stage of tooth development, we show that suboptimal levels of BMP signaling do not lead to abnormal morphogenesis, as suggested by mutants affecting BMP signaling, but to a 24-h stalling of the intrinsic developmental clock of the tooth. During this time, BMP levels accumulate to reach critical levels whereupon tooth development restarts, accelerates to catch up with development of the rest of the embryo and completes normal morphogenesis. This suggests that individual organs can autonomously control their developmental timing to adjust their stage of development to that of other organs. We also find that although BMP signaling is critical for the bud-to-cap transition in all teeth, levels of BMP signaling are regulated differently in multicusped teeth. We identify an interaction between two homeodomain transcription factors, Barx1 and Msx1, which is responsible for setting critical levels of BMP activity in multicusped teeth and provides evidence that correlates the levels of Barx1 transcriptional activity with cuspal complexity. This study highlights the importance of absolute levels of signaling activity for development and illustrates remarkable self-regulation in organogenesis that ensures coordination of developmental processes such that timing is subordinate to developmental structure." | |||
|} | |} | ||
==Human BMP Family== | |||
{{Bmp family table}} | |||
{{Bmp family collapsetable}} | |||
==Structure== | ==Structure== | ||
{| | |||
! colspan=2|BMP Superfamily Canonical Signalling | |||
|- | |||
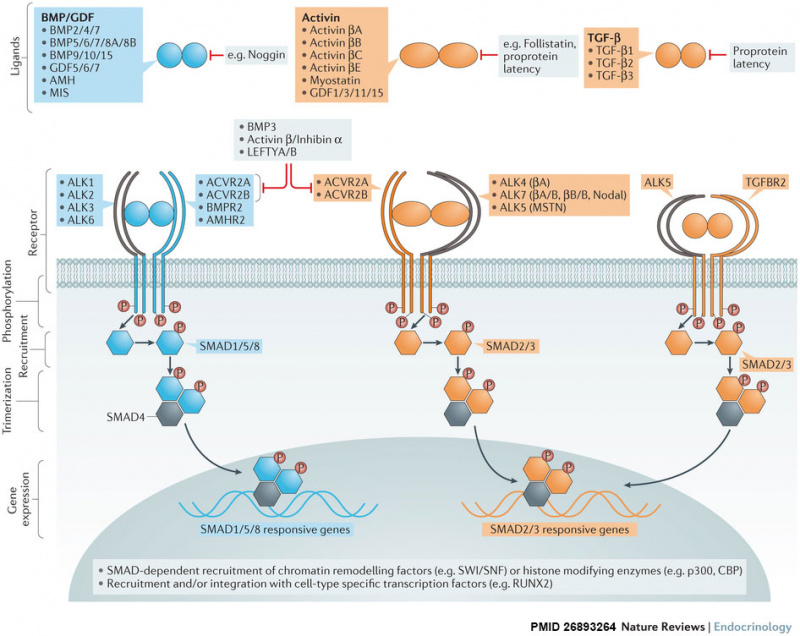
| colspan=2|[[File:BMP superfamily canonical signalling.jpg|800px]] | |||
|- | |||
| Over 30 bone morphogenetic protein (BMP) superfamily ligands have been discovered in humans. Most are secreted as mature disulfide-linked dimers, with the exception of TGF-β1, TGF-β2 and TGF-β3, which can be secreted in a latent form and require proteolytic activation. BMPs signal through a multimeric cell surface complex consisting of two type I receptors and two type II receptors. | |||
Type I and type II BMP receptors are single pass transmembrane proteins with an intracellular serine/threonine kinase domain. After ligand binding, type II receptors phosphorylate (P) the type I receptors. | |||
Activated type I receptors recruit and phosphorylate pathway-specific R-SMADs (SMAD1, SMAD5 and SMAD8 (blue pathway), and SMAD2 and SMAD3 (orange pathway)), which can form trimers with SMAD4 and translocate to the nucleus. SMADs have intrinsic DNA-binding activity and are able to regulate gene expression by recruitment of chromatin-remodelling machinery and integration with tissue-specific transcription factors. SMAD8 is also known as SMAD9. | |||
The pathway can be antagonized by many mechanisms including neutralization of ligands by secreted traps such as noggin or follistatin, secretion of latent ligands bound to their propeptides, or via titration of receptors by nonsignalling ligands such as BMP3, activin β/inhibin α dimers or LEFTY monomers. | |||
| width=300px valign=top| | |||
* ACVR - activin receptor | |||
* ALK - activin receptor-like kinase | |||
* AMH - anti-Müllerian hormone | |||
* AMHR2 - AMH receptor 2 | |||
* BMPR - BMP receptor | |||
* GDF - growth/differentiation factor | |||
* TGF - transforming growth factor | |||
* TGFBR - TGF-β receptor | |||
Figure from recent BMP review.{{#pmid:26893264|PMID26893264}} | |||
|} | |||
===Gene=== | ===Gene=== | ||
* Mouse BMP4 [http://www.ncbi.nlm.nih.gov/gene?term=12159 gene] | [http://www.ncbi.nlm.nih.gov/protein/CAM27178.1 protein] | * Mouse BMP4 [http://www.ncbi.nlm.nih.gov/gene?term=12159 gene] | [http://www.ncbi.nlm.nih.gov/protein/CAM27178.1 protein] | ||
* Human BMP15 gene at Xp11.2 | |||
==Function== | ==Function== | ||
===BMP4=== | |||
{| | {| | ||
| [[File:Mouse Bmp4 expression limb and face 01.jpg|600px]] | | [[File:Mouse Bmp4 expression limb and face 01.jpg|600px]] | ||
Mouse Bmp4 expression limb and face.< | Mouse Bmp4 expression limb and face.{{#pmid:22701669|PMID22701669}} | ||
| | | valign="bottom"|{{Mouse Face Bmp4 movie}} | ||
|} | |||
===Neural Development=== | |||
During gastrulation the BMP pathway is antagonised and involved with neural induction. Neural induction signaling through the BMP-regulated Smad1/5 proteins appears to be controlled by fibroblast growth factor (FGF)-regulated Ca<sup>2+</sup> entry activating calcineurin (CaN) that in turn dephosphorylates Smad1/5 proteins.{{#pmid:24698271|PMID24698271}} | |||
:'''Links:''' {{Neural}} | |||
====Genital Development==== | |||
'''Bmp4 is an essential growth factor for the initiation of {{genita}}l tubercle (GT) outgrowth'''{{#pmid:30714224|PMID30714224}} | |||
:"The external genitalia are appendage organs outgrowing from the posterior body trunk. Murine genital tubercle (GT), anlage of external genitalia, initiates its outgrowth from embryonic day (E) {{ME10.5}} as a bud structure. Several growth factors such as fibroblast growth factor (FGF), Wnt and Sonic hedgehog ({{Shh}}) are essential for the GT outgrowth. However, the mechanisms of initiation of GT outgrowth are poorly understood. We previously identified bone morphogenetic protein ({{Bmp}}) signaling as a negative regulator for GT outgrowth. We show here novel aspects of Bmp4 functions for GT outgrowth. We identified the Bmp4 was already expressed in cloaca region at E9.5, before GT outgrowth. To analyze the function of Bmp4 at early stage for the initiation of GT outgrowth, we utilized the Hoxa3-Cre driver and Bmp4 flox/flox mouse lines. Hoxa3 Cre/+ ; Bmp4 flox/flox mutant mice showed the hypoplasia of GT with reduced expression of outgrowth promoting genes such as Wnt5a, Hoxd13 and p63, whereas Shh expression was not affected. Formation of distal urethral epithelium (DUE) marked by the Fgf8 expression is essential for controlling mesenchymal genes expression in GT and subsequent its outgrowth. Furthermore, Fgf8 expression was dramatically reduced in such mutant mice indicating the defective DUE formation. Hence, current results indicate that Bmp4 is an essential growth factor for the initiation of GT outgrowth independent of Shh signaling. Thus, Bmp4 positively regulates for the formation of DUE. The current study provides new insights into the function of Bmp signaling at early stage for the initiation of GT outgrowth." | |||
:'''Links:''' {{genital}} | |||
===BMP15=== | |||
====Oocyte Development==== | |||
{| | |||
| [[File:Ovarian follicle molecular interactions 01.jpg|300px|alt=Ovarian follicle molecular interactions]] | |||
| [[File:Bovine ovarian follicle BMP15 and GDF9.jpg|300px|alt=Bovine ovarian follicle BMP15 and GDF9]] | |||
|- | |- | ||
| | | Molecular paracrine interactions involving BMP15 signaling{{#pmid:24711992|PMID24711992}} | ||
| | | Localization of BMP15 in calf and cow follicles{{#pmid:21401961|PMID21401961}} | ||
|} | |||
|} | |} | ||
:'''Links:''' [[Oocyte Development]] | |||
===Limb Development=== | |||
Bmp2, Bmp4 and Bmp7 are co-required in the mouse AER for normal digit patterning but not limb outgrowth{{#pmid:22662233|PMID22662233}} | |||
:"Outgrowth and patterning of the vertebrate limb requires a functional apical ectodermal ridge (AER). The AER is a thickening of ectodermal tissue located at the distal end of the limb bud. Loss of this structure, either through genetic or physical manipulations results in truncation of the limb. A number of genes, including Bmps, are expressed in the AER. Previously, it was shown that removal of the BMP receptor Bmpr1a specifically from the AER resulted in complete loss of hindlimbs suggesting that Bmp signaling in the AER is required for limb outgrowth. In this report, we genetically removed the three known AER-expressed Bmp ligands, Bmp2, Bmp4 and Bmp7 from the AER of the limb bud using floxed conditional alleles and the Msx2-cre allele. Surprisingly, only defects in digit patterning and not limb outgrowth were observed. In triple mutants, the anterior and posterior AER was present but loss of the central region of the AER was observed. These data suggest that Bmp ligands expressed in the AER are not required for limb outgrowth but instead play an essential role in maintaining the AER and patterning vertebrate digits." | |||
[[File:Limb AER BMP expression01.jpg|800px]] | |||
:'''Links:''' [[Musculoskeletal_System_-_Limb_Development|Limb Development]] | |||
===Blood Vessel Development=== | |||
BMP/SMAD signaling pathway regulates angiogenic sprouting and is involved in embryo vascular development. | |||
==Signaling Pathway== | ==Signaling Pathway== | ||
Identified BMP modulators:{{#pmid:23573253|PMID23573253}} Noggin, Chordin, Chordin-like 1, Chordin-like 2, Twisted gastrulation, Dan, BMPER, Sost, Sostdc1, Follistatin, Follistatin-like 1, Follistatin-like 5 and Tolloid. | |||
===Receptor=== | ===Receptor=== | ||
===Intracellular Signaling=== | ===Intracellular Signaling=== | ||
===SNW-domain containing protein 1=== | |||
(SNW1, SKI-INTERACTING PROTEIN; SKIIP) | |||
A protein that interacts with nuclear receptors and enhances ligand-activated transcription, also called a nuclear receptor co-activator. | |||
'''Regulator of Spatial BMP Activity, Neural Plate Border Formation, and Neural Crest Specification in Vertebrate Embryos'''{{#pmid:21358802|PMID21358802}} | |||
:"Bone morphogenetic protein (BMP) gradients provide positional information to direct cell fate specification, such as patterning of the vertebrate ectoderm into neural, neural crest, and epidermal tissues, with precise borders segregating these domains. However, little is known about how BMP activity is regulated spatially and temporally during vertebrate development to contribute to embryonic patterning, and more specifically to neural crest formation. Through a large-scale in vivo functional screen in Xenopus for neural crest fate, we identified an essential regulator of BMP activity, SNW1. SNW1 is a nuclear protein known to regulate gene expression. Using antisense morpholinos to deplete SNW1 protein in both Xenopus and zebrafish embryos, we demonstrate that dorsally expressed SNW1 is required for neural crest specification, and this is independent of mesoderm formation and gastrulation morphogenetic movements. By exploiting a combination of immunostaining for phosphorylated Smad1 in Xenopus embryos and a BMP-dependent reporter transgenic zebrafish line, we show that SNW1 regulates a specific domain of BMP activity in the dorsal ectoderm at the neural plate border at post-gastrula stages. We use double in situ hybridizations and immunofluorescence to show how this domain of BMP activity is spatially positioned relative to the neural crest domain and that of SNW1 expression. Further in vivo and in vitro assays using cell culture and tissue explants allow us to conclude that SNW1 acts upstream of the BMP receptors. Finally, we show that the requirement of SNW1 for neural crest specification is through its ability to regulate BMP activity, as we demonstrate that targeted overexpression of BMP to the neural plate border is sufficient to restore neural crest formation in Xenopus SNW1 morphants. We conclude that through its ability to regulate a specific domain of BMP activity in the vertebrate embryo, SNW1 is a critical regulator of neural plate border formation and thus neural crest specification." | |||
==Additional Images== | |||
<gallery> | |||
File:Oocyte BMP15 and GDF9 effects.jpg|Bovine oocyte BMP15 effects PMID 25058588 | |||
File:Mouse_E11.0_Bmp4_01.jpg|Mouse E11.0 body - Bmp4 | |||
File:Mouse_E11.5_Bmp4_01.jpg|Mouse E11.5 body - Bmp4 | |||
File:Mouse_Bmp4_expression_limb_and_face_01.jpg|Mouse E9.5-13.5 face and limb - Bmp4 | |||
File:Mouse_Bmp4_expression_face_01.jpg|Mouse E9.5-13.5 face - Bmp4 | |||
</gallery> | |||
==OMIM== | ==OMIM== | ||
{{Template: | {{About_OMIM}} | ||
:'''Links:''' [http://omim.org/entry/300247 OMIM300247] | |||
==Abnormalities== | |||
===Infertility=== | |||
{| | |||
|+ '''Selected Female Infertility Genes ''' | |||
|-bgcolor="CEDFF2" | |||
! Gene<br>abbreviation | |||
! Name | |||
! Gene Location | |||
! Online Mendelian<br>Inheritance in Man ([https://www.omim.org OMIM]) | |||
! HUGO Gene Nomenclature<br>Committee ([https://www.genenames.org HGNC]) | |||
! GeneCards ([https://www.genecards.org GCID]) | |||
! Diagnosis | |||
|- | |||
| BMP15 || Bone morphogenetic protein 15 || {{ChrX}}p11.22 || [https://www.omim.org/entry/300247 300247] || [https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:1068 1068] || [https://www.genecards.org/cgi-bin/carddisp.pl?gene=GC0XP050910 GC0XP050910] || Primary ovarian insufficiency | |||
|- | |||
| Colspan=7| | |||
|-bgcolor="F5FFFA" | |||
| Colspan=7| Table data source{{#pmid:29199274|PMID29199274}} (table 1) '''Links:''' {{Fertilization}} | {{oocyte}} | {{ovary}} | | [[Template:Female Infertility Genes table 1|Female Infertility Genes]] | {{spermatozoa}} | {{testis}} | [[Template:Male Infertility Genes table 1|Male Infertility Genes]] | [[Abnormal Development - Genetic|Genetic Abnormalities]] | {{ART}}<br> | |||
Primary ovarian insufficiency - depletion or dysfunction of ovarian follicles with cessation of menses before age 40 years. | |||
|} | |||
{{Female Infertility Genes collapse table 1}} | |||
==References== | ==References== | ||
<references/> | <references/> | ||
===Reviews=== | ===Reviews=== | ||
{{#pmid:30340367}} | |||
{{#pmid:26893264}} | |||
{{#pmid:26113727}} | |||
{{#pmid:24174749}} | |||
{{#pmid:22327483}} | |||
{{#pmid:17029022}} | |||
{{#pmid:15621726}} | |||
===Articles=== | ===Articles=== | ||
{{#pmid:24698271}} | |||
{{#pmid:23055827}} | |||
{{#pmid:22693607}} | |||
===Online Textbooks=== | ===Online Textbooks=== | ||
* '''Molecular Biology of the Cell'''. 4th edition. Alberts B, Johnson A, Lewis J, et al.New York: Garland Science; 2002. | * '''Molecular Biology of the Cell'''. 4th edition. Alberts B, Johnson A, Lewis J, et al.New York: Garland Science; 2002. [http://www.ncbi.nlm.nih.gov/books/NBK26822/#A2874 Signal Proteins of the TGF-β Superfamily Act Through Receptor Serine/Threonine Kinases and Smads] | ||
* '''Madame Curie Bioscience Database''' (Internet). Austin (TX): Landes Bioscience; 2000. [http://www.ncbi.nlm.nih.gov/books/NBK6525 TGFβ-dependent Epithelial-Mesenchymal Transition] | |||
* '''Madame Curie Bioscience Database''' | |||
===Search PubMed=== | ===Search PubMed=== | ||
| Line 70: | Line 239: | ||
{{External Links}} | {{External Links}} | ||
* NCBI - [http://www.ncbi.nlm.nih.gov/gene?term=12159 Mouse BMP4 gene] | |||
{{Glossary}} | |||
{{ | {{Footer}} | ||
[[Category:Developmental Signal]] [[Category:BMP]] [[Category:Molecular]] | [[Category:Developmental Signal]] [[Category:BMP]] [[Category:Molecular]] | ||
Revision as of 10:21, 25 June 2019
| Embryology - 25 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
Belongs to the transforming growth factor-beta (TGFB) superfamily, humans have 15 members in this protein signaling family. The proteins are synthesized as prepropeptides, then cleaved, and processed into dimeric proteins.
- TGFB family members: TGFB1, TGFB, TGFB3, bone morphogenetic proteins Bmp-2A, Bmp-2B, Bmp-3, and Bmp-6. mullerian inhibitory substance.
Mouse Bmp4 expression face.[1]
- BMP Mouse Links: Face and limb E9.5-13.5 | Face E9.5-13.5 | Body E11.0 | Body E11.5 | BMP | Mouse Development
Growth Differentiation Factor-6 (Gdf6) is a member of the Bone Morphogenetic Protein (BMP) family of secreted signaling molecules.
| Factor Links: AMH | hCG | BMP | sonic hedgehog | bHLH | HOX | FGF | FOX | Hippo | LIM | Nanog | NGF | Nodal | Notch | PAX | retinoic acid | SIX | Slit2/Robo1 | SOX | TBX | TGF-beta | VEGF | WNT | Category:Molecular |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Bone Morphogenetic Protein | BMP | BMP4 | BMP7 | BMP15
|
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Human BMP Family
| Table - Human Bmp Family | ||||
| Approved Symbol |
Approved Name | Previous Symbols |
Synonyms | Chromosome |
|---|---|---|---|---|
| BMP1 | bone morphogenetic protein 1 | PCOLC | 8p21.3 | |
| BMP2 | bone morphogenetic protein 2 | BMP2A | 20p12.3 | |
| BMP3 | bone morphogenetic protein 3 | 4q21.21 | ||
| BMP4 | bone morphogenetic protein 4 | BMP2B | 14q22.2 | |
| BMP5 | bone morphogenetic protein 5 | 6p12.1 | ||
| BMP6 | bone morphogenetic protein 6 | VGR | VGR1 | 6p24.3 |
| BMP7 | bone morphogenetic protein 7 | OP-1 | 20q13.31 | |
| BMP8A | bone morphogenetic protein 8a | 1p34.3 | ||
| BMP8B | bone morphogenetic protein 8b | BMP8 | OP-2 | 1p34.2 |
| BMP10 | bone morphogenetic protein 10 | 2p13.3 | ||
| BMP15 | bone morphogenetic protein 15 | GDF9B | Xp11.22 | |
| Links: Developmental Signals - Bone Morphogenetic Protein | OMIM BMP2 | HGNC | Bmp Family | Sox Family | Tbx Family | ||||
| Human BMP Family | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Structure
| BMP Superfamily Canonical Signalling | |
|---|---|
| |
| Over 30 bone morphogenetic protein (BMP) superfamily ligands have been discovered in humans. Most are secreted as mature disulfide-linked dimers, with the exception of TGF-β1, TGF-β2 and TGF-β3, which can be secreted in a latent form and require proteolytic activation. BMPs signal through a multimeric cell surface complex consisting of two type I receptors and two type II receptors.
Activated type I receptors recruit and phosphorylate pathway-specific R-SMADs (SMAD1, SMAD5 and SMAD8 (blue pathway), and SMAD2 and SMAD3 (orange pathway)), which can form trimers with SMAD4 and translocate to the nucleus. SMADs have intrinsic DNA-binding activity and are able to regulate gene expression by recruitment of chromatin-remodelling machinery and integration with tissue-specific transcription factors. SMAD8 is also known as SMAD9. The pathway can be antagonized by many mechanisms including neutralization of ligands by secreted traps such as noggin or follistatin, secretion of latent ligands bound to their propeptides, or via titration of receptors by nonsignalling ligands such as BMP3, activin β/inhibin α dimers or LEFTY monomers. |
|
Gene
Function
BMP4
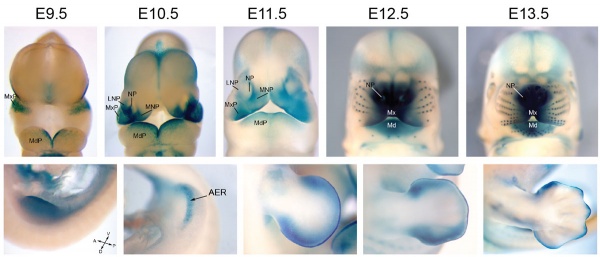
Mouse Bmp4 expression limb and face.[1] |
|
Neural Development
During gastrulation the BMP pathway is antagonised and involved with neural induction. Neural induction signaling through the BMP-regulated Smad1/5 proteins appears to be controlled by fibroblast growth factor (FGF)-regulated Ca2+ entry activating calcineurin (CaN) that in turn dephosphorylates Smad1/5 proteins.[10]
- Links: neural
Genital Development
Bmp4 is an essential growth factor for the initiation of Template:Genital tubercle (GT) outgrowth[3]
- "The external genitalia are appendage organs outgrowing from the posterior body trunk. Murine genital tubercle (GT), anlage of external genitalia, initiates its outgrowth from embryonic day (E) E10.5 as a bud structure. Several growth factors such as fibroblast growth factor (FGF), Wnt and Sonic hedgehog (Shh) are essential for the GT outgrowth. However, the mechanisms of initiation of GT outgrowth are poorly understood. We previously identified bone morphogenetic protein (Bmp) signaling as a negative regulator for GT outgrowth. We show here novel aspects of Bmp4 functions for GT outgrowth. We identified the Bmp4 was already expressed in cloaca region at E9.5, before GT outgrowth. To analyze the function of Bmp4 at early stage for the initiation of GT outgrowth, we utilized the Hoxa3-Cre driver and Bmp4 flox/flox mouse lines. Hoxa3 Cre/+ ; Bmp4 flox/flox mutant mice showed the hypoplasia of GT with reduced expression of outgrowth promoting genes such as Wnt5a, Hoxd13 and p63, whereas Shh expression was not affected. Formation of distal urethral epithelium (DUE) marked by the Fgf8 expression is essential for controlling mesenchymal genes expression in GT and subsequent its outgrowth. Furthermore, Fgf8 expression was dramatically reduced in such mutant mice indicating the defective DUE formation. Hence, current results indicate that Bmp4 is an essential growth factor for the initiation of GT outgrowth independent of Shh signaling. Thus, Bmp4 positively regulates for the formation of DUE. The current study provides new insights into the function of Bmp signaling at early stage for the initiation of GT outgrowth."
- Links: genital
BMP15
Oocyte Development
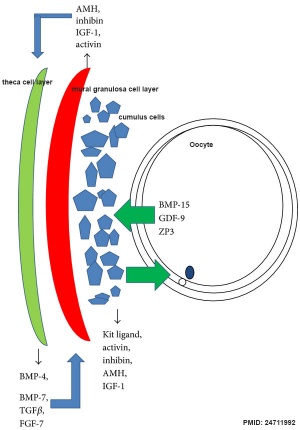
|
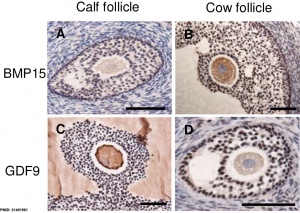
|
| Molecular paracrine interactions involving BMP15 signaling[11] | Localization of BMP15 in calf and cow follicles[12] |
- Links: Oocyte Development
Limb Development
Bmp2, Bmp4 and Bmp7 are co-required in the mouse AER for normal digit patterning but not limb outgrowth[13]
- "Outgrowth and patterning of the vertebrate limb requires a functional apical ectodermal ridge (AER). The AER is a thickening of ectodermal tissue located at the distal end of the limb bud. Loss of this structure, either through genetic or physical manipulations results in truncation of the limb. A number of genes, including Bmps, are expressed in the AER. Previously, it was shown that removal of the BMP receptor Bmpr1a specifically from the AER resulted in complete loss of hindlimbs suggesting that Bmp signaling in the AER is required for limb outgrowth. In this report, we genetically removed the three known AER-expressed Bmp ligands, Bmp2, Bmp4 and Bmp7 from the AER of the limb bud using floxed conditional alleles and the Msx2-cre allele. Surprisingly, only defects in digit patterning and not limb outgrowth were observed. In triple mutants, the anterior and posterior AER was present but loss of the central region of the AER was observed. These data suggest that Bmp ligands expressed in the AER are not required for limb outgrowth but instead play an essential role in maintaining the AER and patterning vertebrate digits."
- Links: Limb Development
Blood Vessel Development
BMP/SMAD signaling pathway regulates angiogenic sprouting and is involved in embryo vascular development.
Signaling Pathway
Identified BMP modulators:[14] Noggin, Chordin, Chordin-like 1, Chordin-like 2, Twisted gastrulation, Dan, BMPER, Sost, Sostdc1, Follistatin, Follistatin-like 1, Follistatin-like 5 and Tolloid.
Receptor
Intracellular Signaling
SNW-domain containing protein 1
(SNW1, SKI-INTERACTING PROTEIN; SKIIP)
A protein that interacts with nuclear receptors and enhances ligand-activated transcription, also called a nuclear receptor co-activator.
Regulator of Spatial BMP Activity, Neural Plate Border Formation, and Neural Crest Specification in Vertebrate Embryos[15]
- "Bone morphogenetic protein (BMP) gradients provide positional information to direct cell fate specification, such as patterning of the vertebrate ectoderm into neural, neural crest, and epidermal tissues, with precise borders segregating these domains. However, little is known about how BMP activity is regulated spatially and temporally during vertebrate development to contribute to embryonic patterning, and more specifically to neural crest formation. Through a large-scale in vivo functional screen in Xenopus for neural crest fate, we identified an essential regulator of BMP activity, SNW1. SNW1 is a nuclear protein known to regulate gene expression. Using antisense morpholinos to deplete SNW1 protein in both Xenopus and zebrafish embryos, we demonstrate that dorsally expressed SNW1 is required for neural crest specification, and this is independent of mesoderm formation and gastrulation morphogenetic movements. By exploiting a combination of immunostaining for phosphorylated Smad1 in Xenopus embryos and a BMP-dependent reporter transgenic zebrafish line, we show that SNW1 regulates a specific domain of BMP activity in the dorsal ectoderm at the neural plate border at post-gastrula stages. We use double in situ hybridizations and immunofluorescence to show how this domain of BMP activity is spatially positioned relative to the neural crest domain and that of SNW1 expression. Further in vivo and in vitro assays using cell culture and tissue explants allow us to conclude that SNW1 acts upstream of the BMP receptors. Finally, we show that the requirement of SNW1 for neural crest specification is through its ability to regulate BMP activity, as we demonstrate that targeted overexpression of BMP to the neural plate border is sufficient to restore neural crest formation in Xenopus SNW1 morphants. We conclude that through its ability to regulate a specific domain of BMP activity in the vertebrate embryo, SNW1 is a critical regulator of neural plate border formation and thus neural crest specification."
Additional Images
OMIM
About OMIM "Online Mendelian Inheritance in Man OMIM is a comprehensive, authoritative, and timely compendium of human genes and genetic phenotypes. The full-text, referenced overviews in OMIM contain information on all known mendelian disorders and over 12,000 genes. OMIM focuses on the relationship between phenotype and genotype. It is updated daily, and the entries contain copious links to other genetics resources." OMIM
- Links: OMIM300247
Abnormalities
Infertility
| Gene abbreviation |
Name | Gene Location | Online Mendelian Inheritance in Man (OMIM) |
HUGO Gene Nomenclature Committee (HGNC) |
GeneCards (GCID) | Diagnosis |
|---|---|---|---|---|---|---|
| BMP15 | Bone morphogenetic protein 15 | Xp11.22 | 300247 | 1068 | GC0XP050910 | Primary ovarian insufficiency |
| Table data source[16] (table 1) Links: fertilization | oocyte | ovary | | Female Infertility Genes | spermatozoa | testis | Male Infertility Genes | Genetic Abnormalities | ART Primary ovarian insufficiency - depletion or dysfunction of ovarian follicles with cessation of menses before age 40 years. | ||||||
| Female Infertility Genes | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
- ↑ 1.0 1.1 Jumlongras D, Lachke SA, O'Connell DJ, Aboukhalil A, Li X, Choe SE, Ho JW, Turbe-Doan A, Robertson EA, Olsen BR, Bulyk ML, Amendt BA & Maas RL. (2012). An evolutionarily conserved enhancer regulates Bmp4 expression in developing incisor and limb bud. PLoS ONE , 7, e38568. PMID: 22701669 DOI.
- ↑ Su YH, Chen YC, Ting HC, Fan TP, Lin CY, Wang KT & Yu JK. (2019). BMP controls dorsoventral and neural patterning in indirect-developing hemichordates providing insight into a possible origin of chordates. Proc. Natl. Acad. Sci. U.S.A. , , . PMID: 31189599 DOI.
- ↑ 3.0 3.1 Kajioka D, Suzuki K, Nakada S, Matsushita S, Miyagawa S, Takeo T, Nakagata N & Yamada G. (2019). Bmp4 is an essential growth factor for the initiation of genital tubercle (GT) outgrowth. Congenit Anom (Kyoto) , , . PMID: 30714224 DOI.
- ↑ Row RH, Pegg A, Kinney B, Farr GH, Maves L, Lowell S, Wilson V & Martin BL. (2018). BMP and FGF signaling interact to pattern mesoderm by controlling basic helix-loop-helix transcription factor activity. Elife , 7, . PMID: 29877796 DOI.
- ↑ Mimura S, Suga M, Okada K, Kinehara M, Nikawa H & Furue MK. (2016). Bone morphogenetic protein 4 promotes craniofacial neural crest induction from human pluripotent stem cells. Int. J. Dev. Biol. , 60, 21-8. PMID: 26934293 DOI.
- ↑ Bier E & De Robertis EM. (2015). EMBRYO DEVELOPMENT. BMP gradients: A paradigm for morphogen-mediated developmental patterning. Science , 348, aaa5838. PMID: 26113727 DOI.
- ↑ Xu PF, Houssin N, Ferri-Lagneau KF, Thisse B & Thisse C. (2014). Construction of a vertebrate embryo from two opposing morphogen gradients. Science , 344, 87-9. PMID: 24700857 DOI.
- ↑ Miletich I, Yu WY, Zhang R, Yang K, Caixeta de Andrade S, Pereira SF, Ohazama A, Mock OB, Buchner G, Sealby J, Webster Z, Zhao M, Bei M & Sharpe PT. (2011). Developmental stalling and organ-autonomous regulation of morphogenesis. Proc. Natl. Acad. Sci. U.S.A. , 108, 19270-5. PMID: 22084104 DOI.
- ↑ Salazar VS, Gamer LW & Rosen V. (2016). BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol , 12, 203-21. PMID: 26893264 DOI.
- ↑ Cho A, Tang Y, Davila J, Deng S, Chen L, Miller E, Wernig M & Graef IA. (2014). Calcineurin signaling regulates neural induction through antagonizing the BMP pathway. Neuron , 82, 109-124. PMID: 24698271 DOI.
- ↑ Chronowska E. (2014). High-throughput analysis of ovarian granulosa cell transcriptome. Biomed Res Int , 2014, 213570. PMID: 24711992 DOI.
- ↑ Hosoe M, Kaneyama K, Ushizawa K, Hayashi KG & Takahashi T. (2011). Quantitative analysis of bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9) gene expression in calf and adult bovine ovaries. Reprod. Biol. Endocrinol. , 9, 33. PMID: 21401961 DOI.
- ↑ Choi KS, Lee C, Maatouk DM & Harfe BD. (2012). Bmp2, Bmp4 and Bmp7 are co-required in the mouse AER for normal digit patterning but not limb outgrowth. PLoS ONE , 7, e37826. PMID: 22662233 DOI.
- ↑ Lorda-Diez CI, Montero JA, Rodriguez-Leon J, Garcia-Porrero JA & Hurle JM. (2013). Expression and functional study of extracellular BMP antagonists during the morphogenesis of the digits and their associated connective tissues. PLoS ONE , 8, e60423. PMID: 23573253 DOI.
- ↑ Wu MY, Ramel MC, Howell M & Hill CS. (2011). SNW1 is a critical regulator of spatial BMP activity, neural plate border formation, and neural crest specification in vertebrate embryos. PLoS Biol. , 9, e1000593. PMID: 21358802 DOI.
- ↑ 16.0 16.1 Harper JC, Aittomäki K, Borry P, Cornel MC, de Wert G, Dondorp W, Geraedts J, Gianaroli L, Ketterson K, Liebaers I, Lundin K, Mertes H, Morris M, Pennings G, Sermon K, Spits C, Soini S, van Montfoort APA, Veiga A, Vermeesch JR, Viville S & Macek M. (2018). Recent developments in genetics and medically assisted reproduction: from research to clinical applications. Eur. J. Hum. Genet. , 26, 12-33. PMID: 29199274 DOI.
Reviews
Schliermann A & Nickel J. (2018). Unraveling the Connection between Fibroblast Growth Factor and Bone Morphogenetic Protein Signaling. Int J Mol Sci , 19, . PMID: 30340367 DOI.
Salazar VS, Gamer LW & Rosen V. (2016). BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol , 12, 203-21. PMID: 26893264 DOI.
Bier E & De Robertis EM. (2015). EMBRYO DEVELOPMENT. BMP gradients: A paradigm for morphogen-mediated developmental patterning. Science , 348, aaa5838. PMID: 26113727 DOI.
Jain AP, Pundir S & Sharma A. (2013). Bone morphogenetic proteins: The anomalous molecules. J Indian Soc Periodontol , 17, 583-6. PMID: 24174749 DOI.
Ruschke K, Hiepen C, Becker J & Knaus P. (2012). BMPs are mediators in tissue crosstalk of the regenerating musculoskeletal system. Cell Tissue Res. , 347, 521-44. PMID: 22327483 DOI.
Gazzerro E & Canalis E. (2006). Bone morphogenetic proteins and their antagonists. Rev Endocr Metab Disord , 7, 51-65. PMID: 17029022 DOI.
Chen D, Zhao M & Mundy GR. (2004). Bone morphogenetic proteins. Growth Factors , 22, 233-41. PMID: 15621726 DOI.
Articles
Cho A, Tang Y, Davila J, Deng S, Chen L, Miller E, Wernig M & Graef IA. (2014). Calcineurin signaling regulates neural induction through antagonizing the BMP pathway. Neuron , 82, 109-124. PMID: 24698271 DOI.
Luo YJ & Su YH. (2012). Opposing nodal and BMP signals regulate left-right asymmetry in the sea urchin larva. PLoS Biol. , 10, e1001402. PMID: 23055827 DOI.
Kuypers E, Collins JJ, Jellema RK, Wolfs TG, Kemp MW, Nitsos I, Pillow JJ, Polglase GR, Newnham JP, Germeraad WT, Kallapur SG, Jobe AH & Kramer BW. (2012). Ovine fetal thymus response to lipopolysaccharide-induced chorioamnionitis and antenatal corticosteroids. PLoS ONE , 7, e38257. PMID: 22693607 DOI.
Online Textbooks
- Molecular Biology of the Cell. 4th edition. Alberts B, Johnson A, Lewis J, et al.New York: Garland Science; 2002. Signal Proteins of the TGF-β Superfamily Act Through Receptor Serine/Threonine Kinases and Smads
- Madame Curie Bioscience Database (Internet). Austin (TX): Landes Bioscience; 2000. TGFβ-dependent Epithelial-Mesenchymal Transition
Search PubMed
- Search PubMed: Bone Morphogenetic Protein
- All Databases: Bone Morphogenetic Protein
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- NCBI - Mouse BMP4 gene
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 25) Embryology Developmental Signals - Bone Morphogenetic Protein. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Developmental_Signals_-_Bone_Morphogenetic_Protein
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G