User talk:Z3291643
Aetiology
Different genetic mechanisms lead to different clinical phenotypes of AS. The most common genetic mechanism leading to AS is the deletion or re-arrangement of maternal chromosome at locus 15q11.2-q13,as the chromosome image shows on the right, accounting for 70% of AS occurrences. [1] This leads to more severe clinical phenotypes of microcephaly, motor difficulties, seizures and impaired speech development. The next most common genetic mechanism is mutation in the UBE3A gene responsible for 10% of AS cases and paternal uniparental disomy and mutation in the imprinting centre (IC), both accounting for 2-5% of AS observed.[2]
AS is caused by the 4 major genetic mechanisms mentioned above and are thus divided into Classes I to IV based on their underlying genetic mechanism. AS patients with the clinical features of AS but no cytogenetic or molecular abnormality in Chromosome 15q11.2-13 are grouped under Class V (summarised in table below). [3]
| Class | Mechanism | Diagnostic Tests | Frequency |
| Ia | De novo deletion | High resolution cytogenetics FISH | 70% [1] |
| Ib | Deletion due to chromosome rearrangement | High resolution cytogenetics FISH | <1% |
| II | Paternal uniparental disomy | RFLP analysis | 2% |
| IIIa | Imprinting defect with IC mutation | Screening of IC for mutations is positive | 2% |
| IIIb | Imprinting defect withouth IC mutation | Screening of IC for mutations is negative | 2% |
| IIIc | Mosaic imprinting defect | Screening of IC for mutations is usually negative | ? |
| IV | UBE3A mutation | Screening of UBE3A for mutations | 5-10% |
| V | No identifiable genetic abnormality | Consider other diagnoses | 5-26% |
Pathogenesis
Chromosome 15 became associated with AS in the 1980s after the observation of many AS patients harboring microdeletions of 15q11.2-15q13. [4] The last decade has shed even more light in the aetiology of AS,as Kishino and Matsuura first identified UBE3A as the causative gene for AS in 1997. [1] [5]
Normal gene product: E6-AP ubiquitin ligase
The function of UBE3A is to encode for E6-AP ubiquitin ligase, however its function in the development of the nervous system and the process of UBE3A mutation leading to cognitive impairment in AS patients is still vague. [6] Past and current research has elucidated that E6-AP ubiquitin ligase is involved in the degradation of four proteins, listed below, but currently, no protein with a direct role in long-term potentiation (LTP) as a target of E6-AP, has been identified. [7] The identification of such target proteins would be a landmark discovery in further understanding of the pathogenesis, aetiology and prospective treatment strategies of AS, as LTP defects is one of the major neurological impairments of AS.
- p53 tumor suppressor protein
- yeast DNA repair protein Rad23
- multicopy maintenance protein 7 subunit
- E6-AP
Abnormal gene product
Disruption to the gene,UBE3A, interferes with the normal functioning of the gene, which is its crucial role in UBE3A ubiquitylation. Problems involving this pathway can lead to various diseases either from the loss or gain of function. [8] In the case of AS, E6-AP is affected, leading to detrimental cognitive impairment in addition to other deficits.[8]
Regional functioning of UBE3A
Despite the uncertain progression of UBE3A mutation and the neurological abnormalities characteristic of AS, animal models have illustrated the vital role of UBE3A in the normal functioning of the brain. This can be exemplified by a study which utilized a mouse model with target inactivation of the gene UBE3A resulting in the manifestation of classical features of AS. [9] In addition, the fact that imprinting of UBE3A takes place only in specific brain regions (cerebellum, olfactory tracts and the hippocampus) confirms the hypothesis that the loss of this gene would be detrimental in the cognitive development. [10] Most genes are inherited in copies of two, one paternal and the other maternal; however, some genes, such as UBE3A only have one functional copy as the other is silenced, coined 'imprinting'. Imprinting of the paternal copy of UBE3A takes place in some cell populations, such as the hippocampal neurons and Purkinje cells in the cerebellum.[11] This means only the maternal UBE3A is functionally expressed in these particular brain regions. Thus a de novo mutation in the maternal copy of the gene will result in the complete absence of functional E6-AP in the associated brain areas. [12]
It is also known that UBE3A plays an important role in synaptic transmission, but exactly how it does so is still not completely understood.[13]
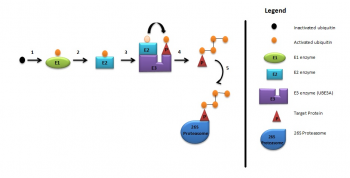
UBE3A ubiquitylation
As briefly mentioned above, UBE3A is a member of the E3 ubiquitin ligase family of enzymes, responsible for the addition of ubiquitin to the target protein for degradation of the ubiquitinated protein, as illustrated in the diagram below. These processes are required for normal human cognitive function.[13] In this way synaptic protein Arc(activity-regulated cytoskeleton-associated protein) is degraded to control synaptic function. Arc is a target protein of UBE3A in dendritic spines found in the hippocampal neurons.[14] Deletion of UBE3A leads to the accumulation of Arc in neurons leading to trafficking of AMPA receptors, resulting in impaired cognitive functions.[6]
UBE3A ubiquitylation consists of:
- Activation of ubiquitin by E1 enzyme
- E1 ezyme transfers ubiquitin to E2 enzyme
- E2 enzyme transfers ubiquitin to UBE3A
- UBE3A attaches activated ubiquitin to target protein which is then polyubiquitylated
- Target protein is then degraded by 26S proteosome.[14]
Phenotype-Genotype correlations
All AS patients show differing extents of cognitive impairment, movement disorder, characteristic behaviours and difficulty in speech and language. [15] However, there seems to be some phenotype-genotype correlations:
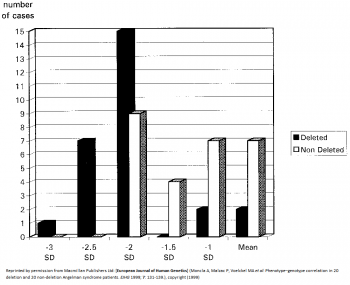
- 5-7Mb deletions result in the most severe phenotypes such as microcephaly, more sever epilepsy and seizures, motor difficulties and language impairment.[16] [17] AS patients with large deletions also present with clinical hypopigmentation, light hair and eye colour due to the close association of OCA2 gene with UBE3A.[18] Individuals under this category have in general no increased BMI [19] This is indicated by the graph on the left, which shows more cases of AS patients with deletions having larger deviations from the mean head circumference measurement than non-deletion AS patients
- AS patients with uniparental disomy (UPD) have better physical growth, fewer motor deficits and lower seizure occurrences [20]
- Individuals with AS resulting from imprinting defects have the least debilitating features, such as higher developmental and language ability than AS caused by other mechanisms. [15]
- The Body Mass Index (BMI) of 33% of the UBE3A mutation patients, and 47–64% of the patients from the classes II (UPD) and III (Imprinting defect) is above the 95th centile [19]
Animal models
- Drosophila
Drosophila is an excellent model to use for better understanding of genetic diseases in humans as they are highly homologous to human UBE3A (hUBE3A), illustrating a high evolutionary conservation. Studies using the Drosophila model have shown that the functional absence of UBE3A resulted in decreased morphogenesis of dendritic branches. This is of interest as dendritic branches cover over 90% of the neuronal surface where synapse between neurons occur. Proper formation and maturation of dendritic spines is also required so they come in contact with other neurons for effective transmission of neuronal signals. Thus proper formation of dendritic branching is pivitol for effective neuronal function and hence cognitive function. [21] Interestingly, overexpression of dUBE3A also gave the same result, consisting of abnormal locomotion and decreased dendritic branching in sensory neurons. [22] This suggests a possible research field for some forms of autism where the region containing UBE3A is duplicated, leading to delayed motor skills and seizures. [23]
- Mice
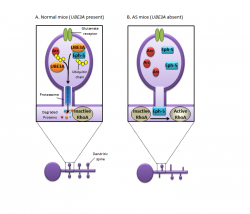
Absence of UBE3A in mice manifest similar phenotype to human AS, as illustrated by motor and learning deficits with inducible seizures. [24] It has been shown through various mice model studies that larger deletions, ie. from UBE3A to GABRB3, more closely mirrors the large 6Mb deletions seen in human AS patients. Numerous tests, such as the Rotarod (for motor coordination and learning ability), Morris water maze test (for spatial learning and memory) and EEG (electroencephalography), were carried out on UBE3A null mutants, which showed the results to be abnormal or impaired. [25] Other studies using mouse models have also outlined the obesity observed in 15% of AS children, albeit it is not the major characterisitics of AS. [26] The diagram on the right clearly highlights the results obtained from such mouse model studies of AS, where the absence of UBE3A accumulated in deficits of motor function and obesity. In addition, excessive drooling and feeding difficulties were also highlighted by mice models with inactivated UBE3A, reflected by defects in fluid consumption and licking. [27]
Pathophysiology
Cognitive defects
AS patients present with abnormally high UBE3A, leading to neurological impairment. Recent identification of key target proteins of Ephexin-58 (Eph-5) and Arc (Activity-Regulated Cytoskeleton-associated protein) have aided in better understanding of the progression of UBE3A to cognitive defects. These two proteins are vital in the understanding of AS as Eph-5 determines synapse numbers and Arc contributes to plasticity and synapse function.[28] Studies utilizing mice models have illustrated that UBE3A null mice could not ubiquitinate Eph-5, thus increasing its levels and limiting the growth of dendritic spines by the activation of RhoA. This subsequent activation of RhoA inhibits synapse formation and results in neuronal defects.
In contrast, mice with normally functioning UBE3A showed phosphorylated Eph-5, resulting from the interaction of two neurons via the activation of EphB signalling receptor. [29] Phosphorylation of Eph-5 allows UBE3A to ubiquitinate and degrade it. This degradation of Eph-5 permits new synaptic connections to form with cells they come into contact with. However, in AS, the absence of UBE3A means Eph-5 cannot be degraded, so RhoA activity is prolonged to reduce synaptic formation.
As mentioned previously, Arc plays a pivitol role in synapse function as it removes synaptic receptors that respond to glutamate, a neurotransmitter. [30] In UBE3A present neurons, UBE3A ubiquitinates Arc, leading to it degradation as illustrated by the diagram on the left. However, in UBE3A deficient neurons, Arc accumulates and prevents the proper synaptic communications essential for normal cognitive functions. [31]
References
- ↑ 1.0 1.1 1.2 <pubmed>8988171</pubmed>
- ↑ <pubmed>19455185</pubmed>
- ↑ <pubmed>10364509</pubmed>
- ↑ <pubmed>15668046</pubmed>
- ↑ <pubmed>8988172</pubmed>
- ↑ 6.0 6.1 <pubmed>20211139</pubmed>
- ↑ <pubmed>12684449</pubmed>
- ↑ 8.0 8.1 <pubmed>9857172</pubmed>
- ↑ <pubmed>11895368</pubmed>
- ↑ <pubmed>20398390</pubmed>
- ↑ <pubmed>9288101</pubmed>
- ↑ <pubmed>9288088</pubmed>
- ↑ 13.0 13.1 <pubmed>9808466</pubmed>
- ↑ 14.0 14.1 <pubmed>20668179</pubmed>
- ↑ 15.0 15.1 <pubmed>11748306</pubmed>
- ↑ <pubmed>9546330</pubmed>
- ↑ <pubmed>20445456</pubmed>
- ↑ <pubmed>16470747</pubmed>
- ↑ 19.0 19.1 <pubmed>15253756</pubmed>
- ↑ <pubmed>16023557</pubmed>
- ↑ <pubmed>18996915</pubmed>
- ↑ <pubmed>18701717</pubmed>
- ↑ <pubmed>16433693</pubmed>
- ↑ <pubmed>21235769</pubmed>
- ↑ <pubmed>20808828</pubmed>
- ↑ <pubmed>17259980</pubmed>
- ↑ <pubmed>18413322</pubmed>
- ↑ <pubmed>21164477</pubmed>
- ↑ <pubmed>21029865</pubmed>
- ↑ <pubmed>17088211</pubmed>
- ↑ <pubmed>20211139</pubmed>