File:Mouse- preimplantation gene expression.jpg
Original file (800 × 612 pixels, file size: 106 KB, MIME type: image/jpeg)
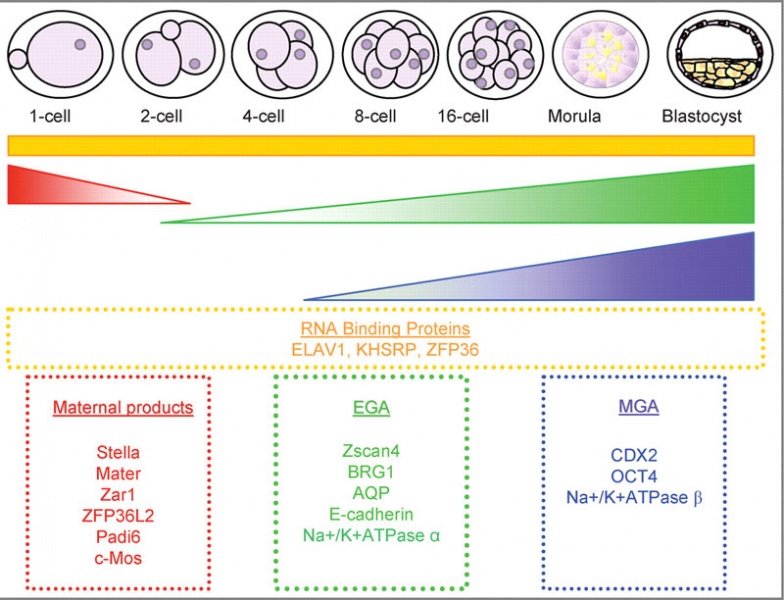
Characterization of global gene expression patterns during preimplantation development
Three groups published stage specific gene array analysis of gene expression in mouse preimplantation embryos in 2004 (Hamatani et al., 2004; Wang et al., 2004; Zeng et al., 2004).
Two main waves of gene expression were observed corresponding with EGA and a second MGA that contributed to events associated with TE differentiation and blastocyst formation (Hamatani et al., 2004). Hamatani et al. (2004), Wang et al. (2004) and Zeng et al. (2004) are landmark studies in the characterization of gene expression patterns during preimplantation development as they have produced extensive catalogues of global stage specific gene expression patterns.
General gene expression patterns are indicated; loss of maternal mRNAs (red); activation of embryonic genome (green); MGA (purple) and continuous expression (orange). Examples of genes expressed in each category are indicated in boxes.
Original file name: Figure 1 F1-1.large.jpg http://molehr.oxfordjournals.org/content/14/12/691/F1.expansion.html
Paper Abstract
- "Preimplantation development shifts from a maternal to embryonic programme rapidly after fertilization. Although the majority of oogenetic products are lost during the maternal to embryonic transition (MET), several do survive this interval to contribute directly to supporting preimplantation development. Embryonic genome activation (EGA) is characterized by the transient expression of several genes that are necessary for MET, and while EGA represents the first major wave of gene expression, a second mid-preimplantation wave of transcription that supports development to the blastocyst stage has been discovered. The application of genomic approaches has greatly assisted in the discovery of stage specific gene expression patterns and the challenge now is to largely define gene function and regulation during preimplantation development. The basic mechanisms controlling compaction, lineage specification and blastocyst formation are defined. The requirement for embryo culture has revealed plasticity in the developmental programme that may exceed the adaptive capacity of the embryo and has fostered important research directions aimed at alleviating culture-induced changes in embryonic programming. New levels of regulation are emerging and greater insight into the roles played by RNA-binding proteins and miRNAs is required. All of this research is relevant due to the necessity to produce healthy preimplantation embryos for embryo transfer, to ensure that assisted reproductive technologies are applied in the most efficient and safest way possible."
Reference
<pubmed>19043080</pubmed>| Mol Hum Reprod.
Special Issue: Emerging Technologies for the Assessment of Gametes and Embryos - The OMICS
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 09:59, 12 October 2010 |  | 800 × 612 (106 KB) | S8600021 (talk | contribs) | ==Characterization of global gene expression patterns during preimplantation development== Three groups published stage specific gene array analysis of gene expression in mouse preimplantation embryos in 2004 (Hamatani et al., 2004; Wang et al., 2004; Ze |
You cannot overwrite this file.
File usage
The following page uses this file: